Abstract
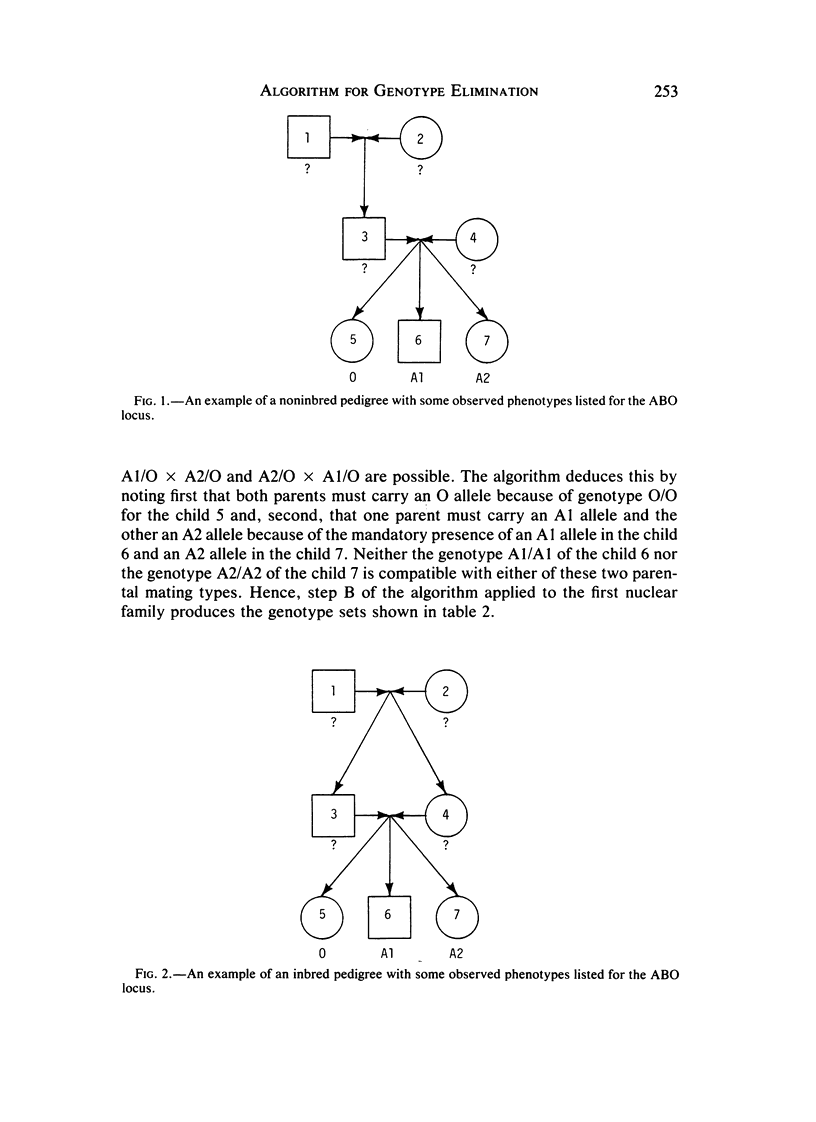
Automatic genotype elimination algorithms for a single locus play a central role in making likelihood computations on human pedigree data feasible. We present a simple algorithm that is fully efficient in pedigrees without loops. This algorithm can be easily coded and has been instrumental in greatly reducing computing times for pedigree analysis. A contrived counter-example demonstrates that some superfluous genotypes cannot be excluded for inbred pedigrees.
Full text
PDF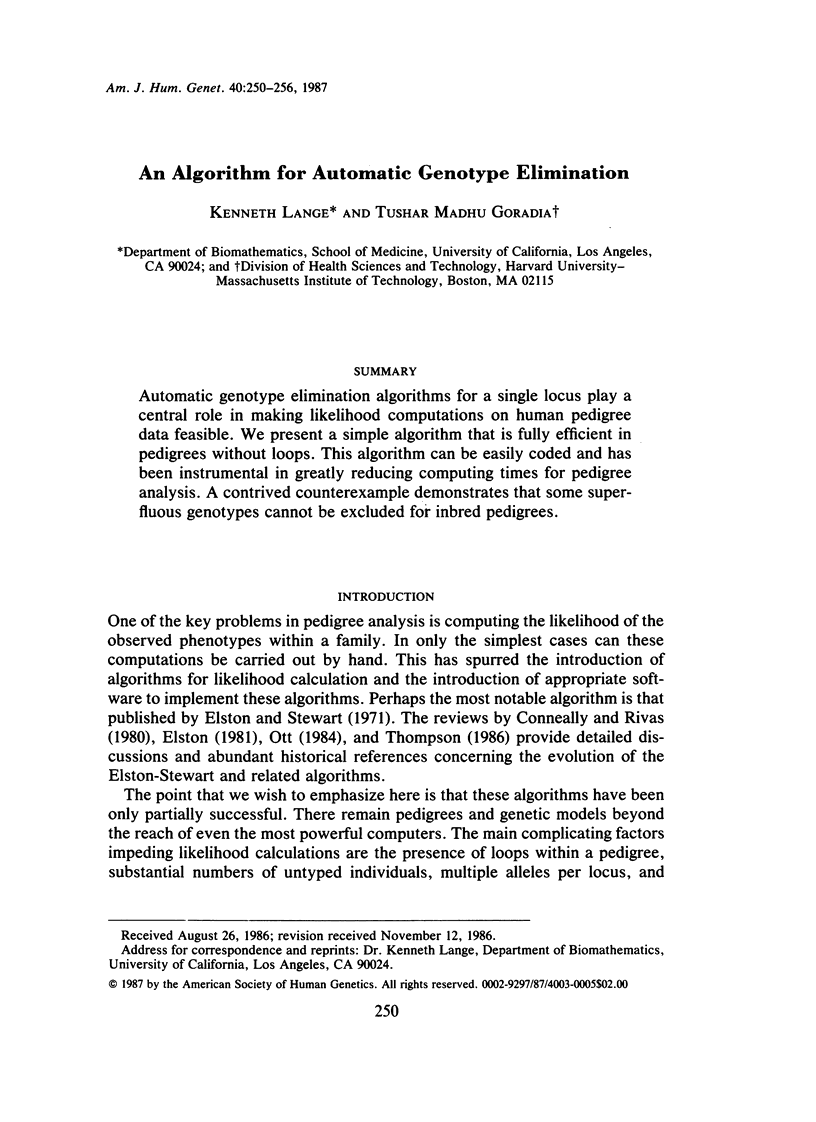





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Conneally P. M., Rivas M. L. Linkage analysis in man. Adv Hum Genet. 1980;10:209–266. doi: 10.1007/978-1-4615-8288-5_3. [DOI] [PubMed] [Google Scholar]
- Elston R. C. Segregation analysis. Adv Hum Genet. 1981;11:63-120, 372-3. doi: 10.1007/978-1-4615-8303-5_2. [DOI] [PubMed] [Google Scholar]
- Elston R. C., Stewart J. A general model for the genetic analysis of pedigree data. Hum Hered. 1971;21(6):523–542. doi: 10.1159/000152448. [DOI] [PubMed] [Google Scholar]
- Lange K., Boehnke M. Extensions to pedigree analysis. V. Optimal calculation of Mendelian likelihoods. Hum Hered. 1983;33(5):291–301. doi: 10.1159/000153393. [DOI] [PubMed] [Google Scholar]


