Abstract
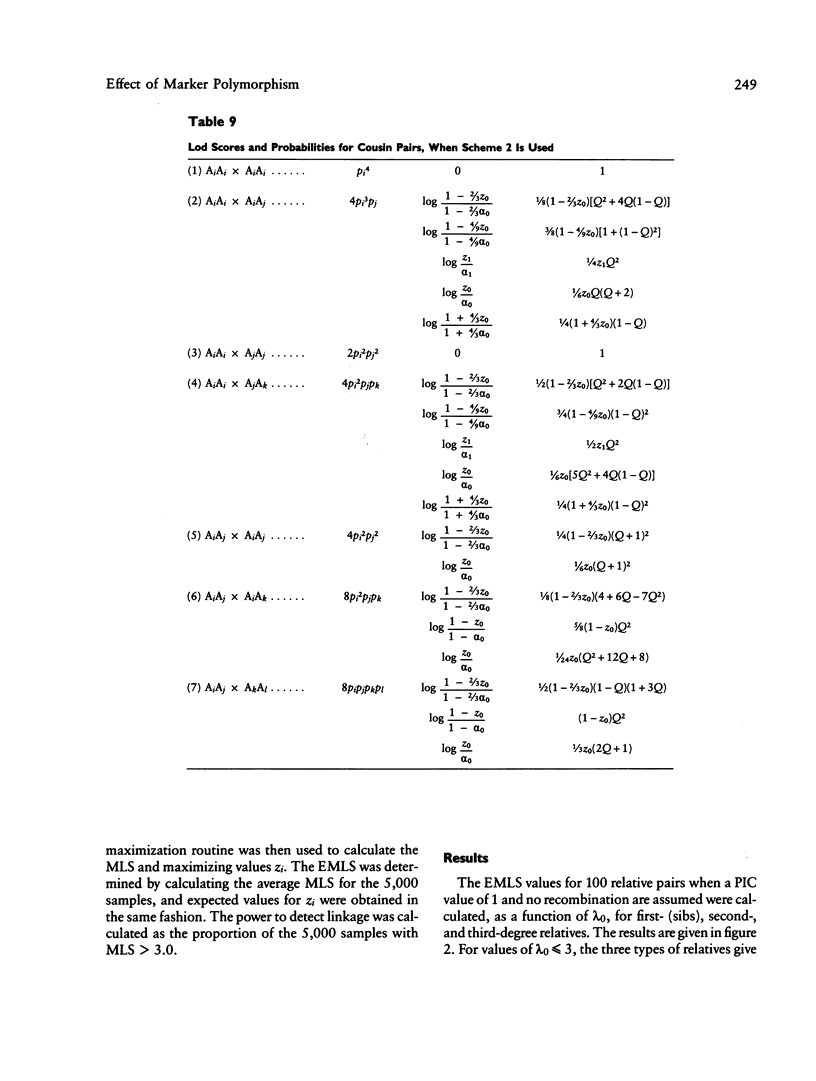
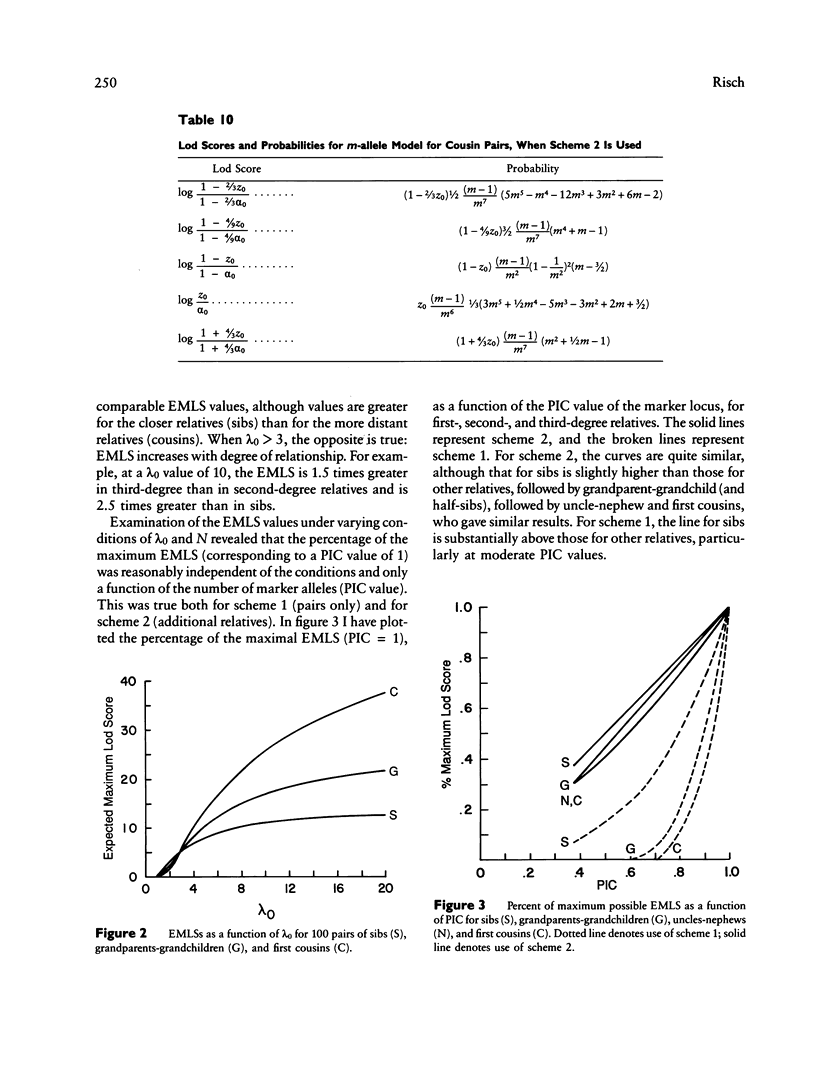
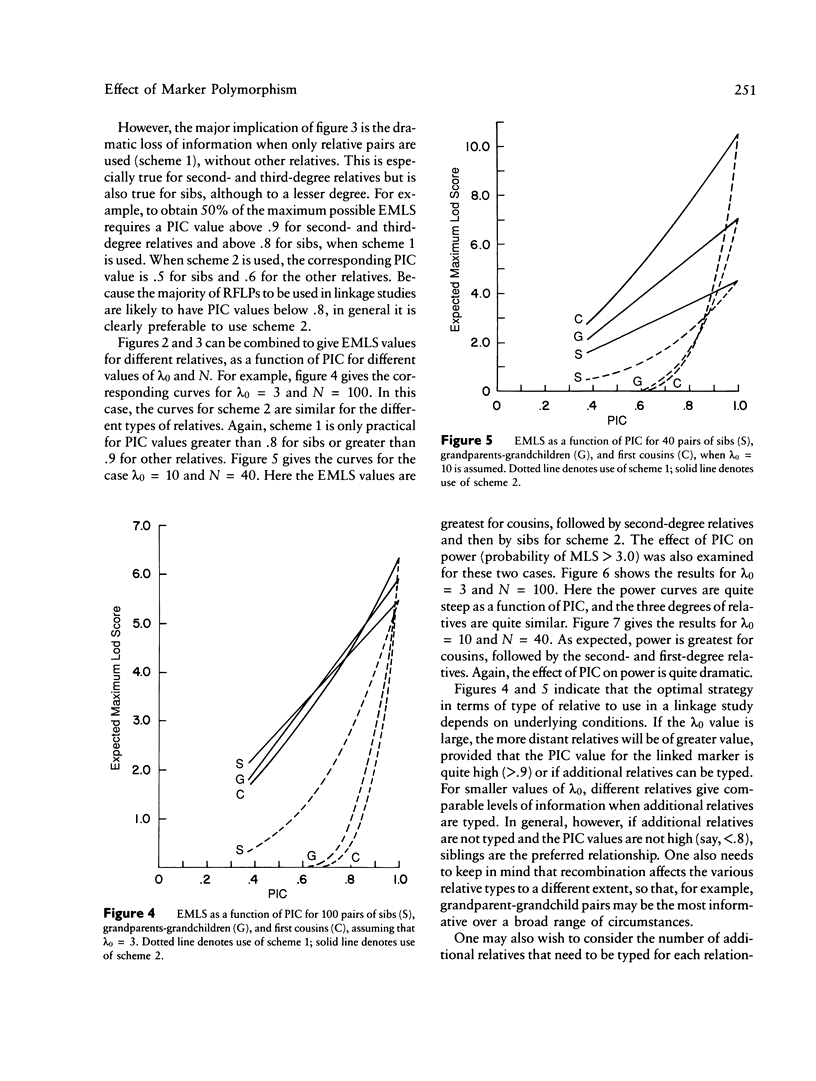
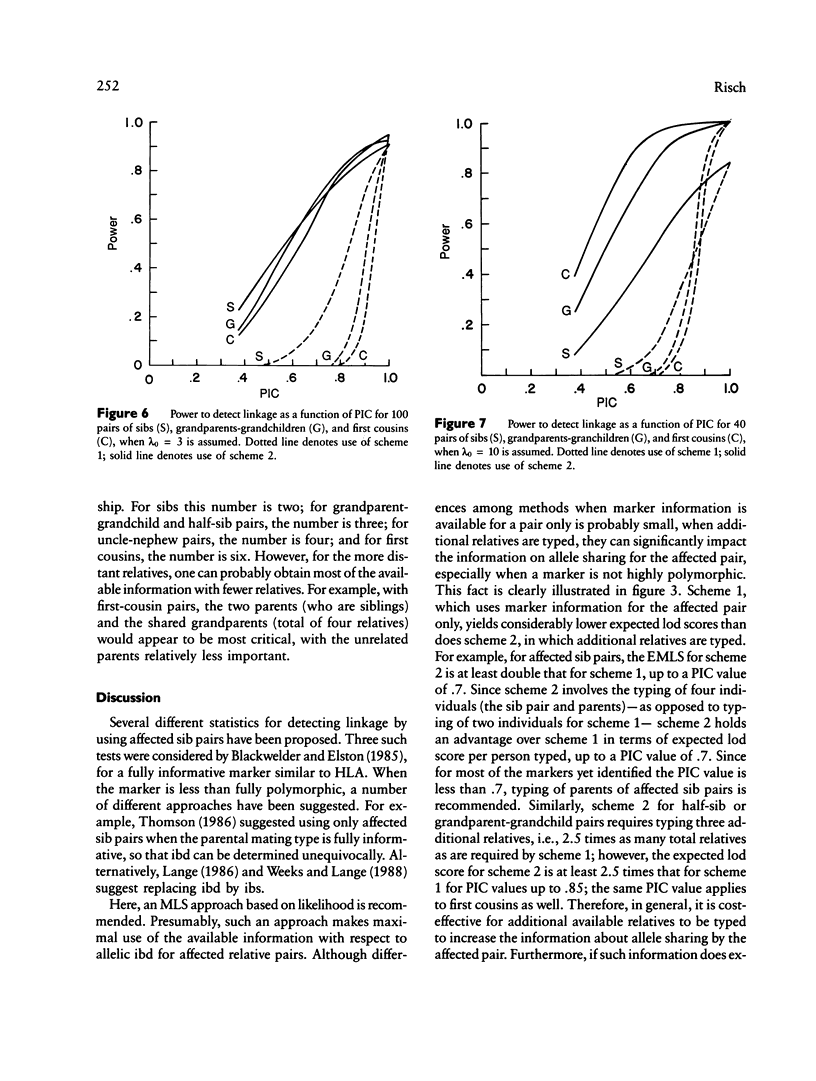
The results from the second paper of this series are reexamined for markers that are not completely polymorphic. A maximum lod score (MLS) criterion is defined for affected relative pairs. The expected MLS (EMLS) is calculated as a function of the marker polymorphic information content (PIC) for various values of lambda R (relative risk ratio) and different relative types by using simulations. An m-allele model with equal allele frequencies is employed. The EMLS is calculated for two sampling strategies: scheme 1, which uses pairs only, and scheme 2, which also includes additional informative relatives. For scheme 2, the percent of the maximum achievable EMLS (i.e., for a marker with a PIC of 1.0) is approximately equal to the marker PIC value for all relative types. For scheme 1, the EMLS is greatly diminished unless PIC is high, especially for distant relatives. For example, scheme 1 is not cost-effective for sibs unless PIC greater than .7; for second- and third-degree relatives, PIC must be greater than .85. Therefore, in general, it will be worthwhile to type additional relatives in linkage studies using affected pairs. The comparative value of sibs versus distant relatives depends on lambda R, recombination theta, and PIC. For large lambda R and PIC values, distant relatives are preferred. Alternatively, for smaller lambda R and PIC values, sibs are best.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blackwelder W. C., Elston R. C. A comparison of sib-pair linkage tests for disease susceptibility loci. Genet Epidemiol. 1985;2(1):85–97. doi: 10.1002/gepi.1370020109. [DOI] [PubMed] [Google Scholar]
- Botstein D., White R. L., Skolnick M., Davis R. W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980 May;32(3):314–331. [PMC free article] [PubMed] [Google Scholar]
- Lange K. The affected sib-pair method using identity by state relations. Am J Hum Genet. 1986 Jul;39(1):148–150. [PMC free article] [PubMed] [Google Scholar]
- MORTON N. E. Sequential tests for the detection of linkage. Am J Hum Genet. 1955 Sep;7(3):277–318. [PMC free article] [PubMed] [Google Scholar]
- Risch N. Genetics of IDDM: evidence for complex inheritance with HLA. Genet Epidemiol. 1989;6(1):143–148. doi: 10.1002/gepi.1370060127. [DOI] [PubMed] [Google Scholar]
- Thomson G. Determining the mode of inheritance of RFLP-associated diseases using the affected sib-pair method. Am J Hum Genet. 1986 Aug;39(2):207–221. [PMC free article] [PubMed] [Google Scholar]
- Weeks D. E., Lange K. The affected-pedigree-member method of linkage analysis. Am J Hum Genet. 1988 Feb;42(2):315–326. [PMC free article] [PubMed] [Google Scholar]


