Abstract
An increasing number of studies suggest the importance of antibodies in the pathogenesis of most systemic and organ-specific autoimmune diseases, although there is considerable controversy over the precise role of the autoantibodies involved. In humans, a major obstacle to progress is the identification and cloning of the relevant autoantibodies and autoantigens. Here, an approach based on the sequential use of antibody phage display and antigen expression libraries is developed and applied to a donor suffering from rheumatoid arthritis (RA), splenomegaly, and peripheral destruction of neutrophils leading to neutropenia (Felty's syndrome). An antibody phage display library was constructed from bone marrow from the donor and a high-affinity human mAb, ANA15, selected by panning against fresh neutrophils and independently by panning against a fixed cell line. The antibody showed strong staining of neutrophils and a number of cell lines. Probing of a λgt11 expression library from an induced myelomonocytic cell line with the mAb ANA15 identified the eukaryotic elongation factor 1A-1 (eEF1A-1) as a novel autoantigen. The specificity of ANA15 was confirmed by reactivity with both purified and recombinant eEF1A-1. Screening of a large panel of sera revealed that 66% of patients with Felty's syndrome had elevated levels of anti-eEF1A-1 antibodies. The cloning of this antibody–antigen pair should permit rational evaluation of any pathogenicity resulting from the interaction and its significance in neutropenia.
Keywords: antibody phage display, cDNA expression libraries, autoimmune neutropenia, human mAb, elongation factor 1A
Autoantibodies, generated as part of an antigen-driven, T cell-dependent response directed to a variety of cellular antigens, are a feature of systemic rheumatic diseases and organ-specific autoimmune diseases. Certain autoantibodies such as anti-DNA antibodies in systemic lupus erythematosus (SLE), antineutrophil cytoplasmic antibodies in systemic vasculitis and antiphospholipid antibodies in antiphospholipid syndrome, are used for differential diagnoses and to measure disease activity and prognosis (1, 2). Such antibodies have often been viewed as a consequence rather than a contributing factor to disease. However, an increasing body of evidence both from clinical studies and animal models in which autoimmune disease occurs spontaneously indicates that antibodies such as anti-DNA in SLE nephritis and antiphospholipid antibodies in antiphospholipid syndrome play an important role in the disease pathogenesis (3–6). In other autoimmune diseases, analysis of animal models has also indicated that the binding of autoantibodies to the corresponding autoantigen may cause or contribute to the inflammatory/fibrogenic response. One recent study suggesting a direct pathogenic role for antibody involves an autoimmune T cell receptor transgenic mouse model. The study showed that disease was conferred by broad autoreactivity, through which T cells stimulated B cells to produce arthritogenic antibodies. These IgG antibodies, directed against an intracellular antigen, were then sufficient to produce the disease, even in the complete absence of any lymphocytes (7, 8).
Recently, studies have also shown that autoantibodies can be used as probes to provide essential clues to the disease initiation process. For example, apoptosis-induced protease reactions in SLE patients have been shown to uniquely fragment autoantigens, exposing novel epitopes to the immune system (9, 10). T and B cells with specificity for those cryptic neoepitopes are not deleted, allowing an immune response to develop.
A general problem in the study of human autoimmunity is the difficulty of identifying relevant autoantigens. Although several autoantigens have been identified by serology, such analysis has several limitations. The use of patient sera as probes for the analysis of recombinant cDNA expression libraries (or peptide libraries), referred to as SEREX, has been described for the identification of novel autoantigens (11–13). However, the use of whole sera is less than straightforward for a number of reasons, including the presence of naturally occurring serum antibodies against bacterial or phage antigens that may complicate antigen selection from bacteriophage expression libraries (12). Furthermore, in the SEREX approach, the serum titer against the antigen of interest should be high, the sera should be relatively monospecific, and large quantities of serum from one time point are required because the serum response may change over time. Finally, the composition of the cDNA expression library may also affect the outcome, because the presence of B cells in the tissue used for construction of the cDNA library may result in incorporation of IgG cDNA in the library. Expression of these fragments may be detected as false-positive clones by the secondary anti-IgG antibody.
An alternative approach to the use of sera in SEREX is to generate an antibody phage display library reflecting the serum response of the donor and subsequently rescue specific mAbs by selection against the target cells or tissue to which the antibody response is directed. Isolated phage antibodies can then be used as probes to screen cDNA expression libraries for isolation of cDNA clones that encode for the corresponding antigen. This approach may abrogate the problem of serum monoreactivity and availability, and of interfering antibacterial and phage antibodies. Multiple serum specificities should readily be dissected, and the individual selected and purified recombinant antibodies used for isolation of cDNA clones encoding for the corresponding antigen. Advantage is taken of the antibody phage display technology that has been shown to be a powerful method for dissecting antibody responses (14–16). However, although multiple human mAbs against a wide range of known antigens have been generated with this technology, there has been limited success in generating human mAbs against novel antigens (17). Recent results on library selection on cell and tissue sections from a number of laboratories including our own, as in part presented here, seem to indicate that these difficulties can be overcome (17, 18).
Felty's syndrome, an extraarticular manifestation of rheumatoid arthritis (RA) involving peripheral destruction of neutrophils resulting in neutropenia, is a complex disease (19–24). Several studies have demonstrated the presence of antineutrophil antibodies in the serum of patients with the syndrome (19–22). In addition, immune complexes have been observed in some of the patients (23, 24) and T cell-mediated suppression of the normal bone marrow granulopoiesis observed in others (23). In contrast to other hematologic autoimmune disorders, there is limited information regarding the antigenic specificities of antineutrophil autoantibodies. Only a few target antigens on neutrophils have been identified, most of which are also recognized by alloimmune sera. Herein, we describe the identification and cloning of a novel autoantigen–autoantibody pair from a patient with Felty's syndrome. From a human antibody library generated from the bone marrow of a donor with this disease, we isolated an autoantibody reactive with neutrophils. This autoantibody was characterized and used as a probe for screening a cDNA expression library, leading to the identification of eukaryotic elongation factor 1A-1 (eEF1A-1), an essential component of the eukaryotic translational apparatus, as a novel autoantigen. The presence of a serum response against purified eEF1A-1 was subsequently demonstrated in 41 of 62 patients with Felty's syndrome. The human antibody and antigen identified provide valuable tools for investigation of the association of anti-eEF1A-1 activity with neutropenia.
Materials and Methods
Library Construction and Phage Selection.
Preparation of RNA from bone marrow lymphocytes and subsequent construction of IgG1 κ/λ Fab libraries using the pComb3 M13 surface display system has been described (15, 25). For antibody selection, the phage libraries generated from the donor with Felty's syndrome were panned separately against fresh isolated neutrophils in suspension and acetone-fixed HEp-2 cells attached to glass slides [antinuclear antigen (ANA) test, Bion Enterprises, Park Ridge, IL]. Neutrophils were prepared from freshly drawn heparinized whole blood by Ficoll-Paque separation and purified by NaCl lysis. Subsequently, the neutrophil pellet was washed twice with PBS and resuspended to ≈5 × 107 cells/ml in PBS. Fab displaying phage resuspended in PBS containing 1% BSA were added to the microtube containing neutrophils and to HEp-2 cells attached to a glass side and incubated for 1 h at 4°C or 24°C, respectively. Unbound phage were removed by washing 4–10 times with PBS containing 1% BSA. Bound phage, enriched for those bearing antigen-binding surface Fabs, were eluted with 0.2 M glycine⋅HCl buffer pH 2.2. The eluted phage were amplified by infection of Escherichia coli and superinfection with M13 helper phage.
Purification of Fabs and ELISA Analysis.
Fab ANA15 was purified from bacterial supernatants by affinity chromatography (16). To assess specificity, supernatants were screened against neutrophils and HEp-2 cells by immunofluorescence (IF) as described below and by ELISA against recombinant eEF1A, rabbit eEF1A purified from reticulocytes (a kind gift from Dr. W. C. Merrick, Case Western Reserve University, Cleveland, OH) and a panel of unrelated antigens, including ovalbumin (Sigma), HIV-1 gp120 (Intracel, Issaquah, WA), and RNA. Human Fabs or rabbit anti-human eEF1A antibody (a kind gift from Dr. G. M. Janssen, University of Leiden, Leiden, The Netherlands) were incubated with the test antigen for 1.5 h at 37°C, followed by washing with PBS/0.05% Tween 20. Detection of bound human Fabs and rabbit antibody was performed with alkaline phosphatase-labeled goat anti-human IgG F(ab′)2 antibody (Pierce) or alkaline phosphatase-labeled goat anti-rabbit IgG antibody (Pierce), and visualized with nitrophenol substrate (Sigma) by reading absorbance at 405 nm.
Sera from patients diagnosed with Felty's syndrome, RA without Felty's syndrome, and SLE (kindly provided by Dr. P. Davis, University of Alberta, Edmonton, Canada; Dr. F. C. Breedveld, Leiden University Hospital, Leiden, The Netherlands; and Dr. R. Fox, Scripps Green Hospital, La Jolla, CA) and 22 healthy normal volunteers were tested for binding to purified eEF1A. The Felty's syndrome patients had RA and spontaneous sustained neutropenia of <2.0 × 109/liter. The neutropenia could not be attributed to any other disease or drug therapy. The diagnoses RA and SLE were defined according to the classification criteria of the American College of Rheumatology. Clinical and laboratory data of most of the patients with Felty's syndrome have been reported (20, 22). The sera, dilutions from 1:800–1:50, were incubated with the test antigen for 1.5 h at 37°C, washed with PBS/0.05% Tween 20. Bound antibody was detected with alkaline phosphatase-labeled F(ab′)2 fragment of goat anti-human IgG Fc-specific antibody (Jackson ImmunoResearch; 1:1,000) and visualized with nitrophenol substrate substrate. Testing for significant differences between means was performed with the unpaired Student t test.
cDNA Cloning and Expression of the 247-aa C-Terminal Domain of Human eEF1A-1.
A λgt11 cDNA library, generated from mRNA from dibutyryl cyclic AMP-induced HL-60 cells (kindly provided by Dr. R. Ye, Scripps Research Institute), was mixed with E. coli Y1090r− cells, and plated onto LB plates. The expressed proteins were transferred to immobilon filters, blocked, and incubated with Fab ANA15 (10 μg/ml) for 1 h. The filters were washed, incubated with horseradish peroxidase-labeled goat anti-human Fab (Pierce; 1:1,500) for 30 min and bound antibody visualized with chemiluminescence-enhancing solution (Pierce) and autoradiography. Plaques corresponding to positively stained points on the filters were subcloned twice. DNA from plate lysates of the positive plaques was isolated by using Lambda TRAP PLUS (CLONTECH) according to the manufacturer's guidelines. The inserts were amplified by PCR and sequenced. The obtained sequences were compared with reported sequences from GenBank.
To avoid expression of λgt11 vector sequences and place the isolated λgt11 cDNA insert into the correct reading frame, a second set of PCR primers were designed and the PCR product cloned into the pBAD TOPO TA vector according to the manufacturer's instructions. The His-tagged eEF1A-1 fragment was purified from the bacterial supernatant by incubation with Nickel beads [Ni-NTA Superflow (Qiagen, Hilden, Germany)]. Supernatant, effluent, wash, and eluate were examined by ELISA and SDS/PAGE by using 10% gels and silver staining.
Indirect Immunofluorescence Analysis and Confocal Laser Scanning Microscopy.
Mammalian cell lines HEp-2, HL-60, Cos, and MB157 were grown in medium containing 10% FBS and allowed to adhere to chambered cover slips (Nunc) for 48 h at 37°C, 5% CO2, to form monolayers. Glass slides with HEp-2 cells attached (ANA-screen, Immuno Concepts, Sacramento, CA or Bion Enterprises) were also used. Cells were incubated with propidium iodine (Sigma) or the following antibodies: purified Fab ANA15 (2–5 μg/ml), donor serum (1:640), preparation bacterial Fab supernatant (1:2), rabbit anti-human serum (1:100), mouse anti-human rough endoplasmic reticulum antibody (1:50; Dako), mouse anti-human nucleoli antibody (1:2; Biodesign International, Kennebunkport, ME), and mouse anti-human α-tubulin antibody (1:100). The cells were washed with PBS and incubated with FITC-labeled (Fab′)2 goat anti-human IgG (Fab′)2 antibody, Texas Red-labeled goat anti-mouse IgG antibody, or CY-5-labeled goat anti-rabbit IgG antibody (all from Jackson ImmunoResearch). The cells were washed with PBS for 15 min and the antifading reagent Slow Fade in PBS/glycerol (Molecular Probes) was added. As a control, all experiments were performed omitting the primary antibody.
Surface Plasmon Resonance.
The kinetics of Fab ANA15 binding to the recombinant eEF1A-1 fragment were determined by surface plasmon resonance-based measurements by using the BIAcore instrument (Amersham Pharmacia). Fab ANA15 (50 μl at a concentration of 5 μg/ml in acetate buffer, pH 5.5) was coupled to a CM5 sensor. The association and dissociation rate constants, kon and koff, were determined under a continuous flow rate of 10 μl/min by using a range of concentrations (31–500 nM) of the recombinant eEF1A-1 fragment, as described (26).
Results
Antibody Selection.
Interest in the role of antibodies in the peripheral destruction of neutrophils observed in patients with Felty's syndrome led us to attempt to identify novel disease-associated antibody–antigen pairs in such patients. Initially, we examined a panel of sera from patients with Felty's syndrome for ANA antibodies, a diagnostic analysis commonly used in the clinic. One of the sera contained IgG antibodies that exhibited an uncharacteristic ANA staining pattern of HEp-2 cells, as well as neutrophil-specific IgG antibodies, as demonstrated by strong binding to neutrophils but no binding to lymphocytes by IF, a second common diagnostic analysis.
To identify and clone the autoantibody–autoantigen pair involved, an IgG/κ/λ Fab phage display library of ≈6 × 106 members was constructed from the bone marrow RNA of the patient. The antibody library was selected against fresh isolated neutrophils in suspension and against acetone-fixed HEp-2 cells attached to glass slides. After four rounds of biopanning, a 40-fold amplification in eluted phage was observed in both selection protocols, indicating enrichment for antigen-binding clones. Phagemid DNA was prepared from the last round of both protocols and the gene III fragment was removed by treatment with the enzymes NheI and SpeI, followed by religation. The reconstructed phagemid was used to transform XL1-Blue cells to produce clones secreting soluble Fab fragments. Subsequent screening of Fab supernates from 25 randomly chosen clones from the HEp-2 panning against acetone-fixed HEp-2 cells attached to glass slides yielded one antibody Fab fragment, ANA15, which exhibited strong binding to the nucleus of HEp-2 cells. Fab supernates from 20 randomly chosen clones from panning against neutrophils were screened against ethanol-fixed neutrophils attached to glass slides and yielded one Fab that bound strongly to neutrophils. The two Fabs obtained by the two different approaches were identical as determined by sequencing of the variable heavy and light chain domains.
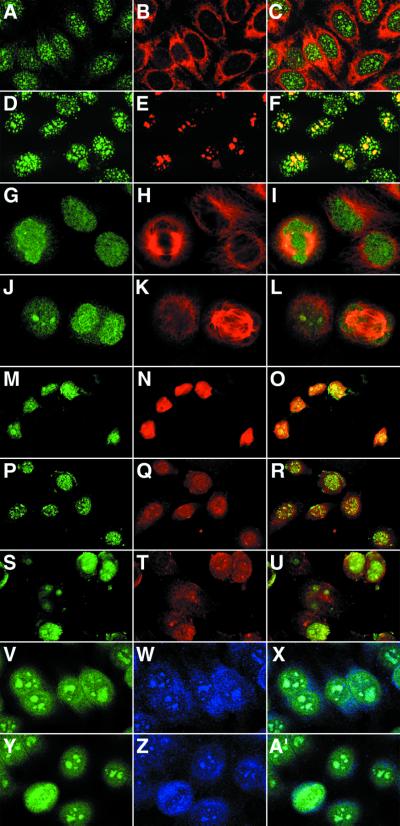
Confocal microscopy of HEp-2 cells demonstrated that Fab ANA15 exhibited a speckled staining pattern exclusively in the nucleus and distinct from the reactivity of previously described ANAs (Figs. 1 and 2). No staining was observed in the cytoplasm, as demonstrated by double staining with a mouse anti-human endoplasmic reticulum antibody (Fig. 1 A–C). In resting cells, the speckles were uneven in size and shape, with some being clumpy as seen with antinucleoli antibodies. However, the number of speckles exceeded that seen with antinucleoli antibodies, and the speckles were localized both in and outside the nucleoli, as demonstrated by double staining with a mouse antinucleolar antibody (Fig. 1 D–F). Fab ANA15 was also examined for binding to ethanol-fixed neutrophils and lymphocytes by IF analysis (Fig. 2). An intense staining at the periphery of the nucleus of the neutrophils was observed. However, surprisingly, no staining of lymphocytes was observed even at high Fab concentration (10 μg/ml). In addition, Fab ANA15 was tested for binding to a panel of transformed cell lines. As shown in Fig. 1 M–U, Fab ANA15 gave nuclear staining of dbcAMP-induced HL-60, Cos, and MDA-MB-157 human breast cancer cells. During cell division (Fig. 1 G–L and Y–A′), where the nuclear membrane was partially disrupted, the staining of HEp-2 cells with Fab ANA15 was more diffuse.
Figure 1.
Cell distribution of eEF1A-1 in different cell types and at different stages of the cell cycle probed by Fab ANA15 with laser scanning confocal microscopy. Mammalian cell lines [HEp-2 (A–L and V–A′); HL-60 (M–O); Cos (P–R); and MB157 (S–U)] were fixed, permeabilized, and stained with Fab ANA15 (all images in the first column) or mouse anti-human endoplasmic reticulum antibody (B), mouse anti-human nucleoli antibody (E), mouse anti-human α-tubulin antibody (H, K), propidium iodine (Red, N, Q, and T), or polyclonal rabbit anti-human eEF1A (W, Z). The images in column 3 are a superposition of columns 1 and 2. Cell distribution of eEF1A-1 in HEp-2 cells during metaphase (G–I), and anaphase (J–L) is also shown. Fab ANA15 was detected with FITC-anti-human IgG(Fab′)2 (green), the mouse antibodies with Texas Red-anti-mouse IgG (red), and the rabbit antibody with CY-5-anti-rabbit IgG antibody (blue).
Figure 2.
Comparison of the staining patterns of Fab ANA15 and the donor serum (patient with Felty's syndrome) on ethanol-fixed HEp-2 cells and neutrophils with indirect IF. For HEp-2 cells, distinctly different staining for the two reagents was observed, whereas similar staining patterns were observed for neutrophils. Fab ANA15 stained neutrophils (arrows) but not neighboring lymphocytes (arrowheads). No staining was observed with a control Fab against HIV-1 gp120 or secondary antibody alone (not shown).
Antigen Selection Using the Recombinant Antibody as Probe.
Because the comparative analysis with sera or antibodies to the known autoantigens indicated that Fab ANA15 recognized a novel autoantigen, we decided to clone the antigen. By using Fab ANA15 as a probe, a cDNA library generated from dbcAMP-induced HL-60 cells expressed in λgt11 was screened. This library was chosen because previous analysis had shown that Fab ANA15 bound to dbcAMP-induced HL-60 cells. After three subcloning cycles, DNA was prepared from Fab ANA15-binding clones and the vector insert amplified by PCR. The PCR amplification of two clones resulted in a 1,025-bp fragment. Sequencing of these two fragments and subsequent comparison with known sequences in GenBank revealed that the insert had 100% identity with 741 nucleotides of the gene encoding human eukaryotic elongation factor 1 alpha (eEF1A-1). Two other clones gave fragments of 400 bp, but the sequences of the inserts showed homology only with bacterial proteins.
The eEF1A-1 insert, omitting adjacent λgt11 vector sequences and placed in the right reading frame, was cloned into the pBAD-TOPO TA vector and the 247-aa C-terminal domain of human eEF1A-1 (aa 216–462) expressed in the LMG194 bacterial strain. The eEF1A-1 protein fragment was purified by using the attached His tag, and when examined by SDS/PAGE, showed a single band of a molecular weight of 28 kDa (Fig. 3A). The supernatant, flow-through, wash, and eluate were further coated onto ELISA wells for subsequent incubation with Fab ANA15. Strong binding of ANA15 was observed with the eluate, whereas no binding was obtained with the flow-through or wash fractions, demonstrating the specificity of ANA15 for the C-terminal domain of human eEF1A-1 (Fig. 3B).
Figure 3.
(A) SDS/PAGE analysis of fractions obtained during purification of cloned recombinant eEF1A-1 fragment. Eluates (1–3), flow-through (FT), and marker (M). The gel shows that the eluted eEF1A-1 fragment has a molecular weight of ≈28 kDa. (B) Analysis of binding of Fab ANA15, polyclonal rabbit anti-human eEF1A-1 antibody, and anti-gp120 Fab L17 (negative control) to the different purification fractions (as shown in A) by using ELISA. Strong reactivity of Fab ANA15 and rabbit anti-human eEF1A-1 with the purified eEF1A-1 fragment, but no reactivity with the flow-through was observed. No binding of Fab L17 to any of the fractions was observed.
Confirming the Specificity of the Autoantibody–Antigen Pair.
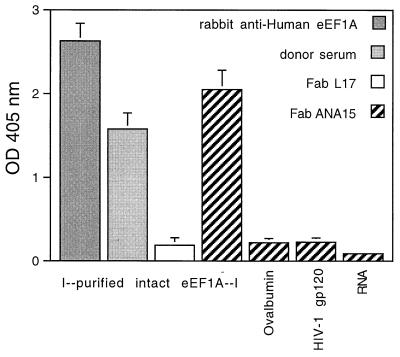
To investigate the interaction of Fab ANA15 and eEF1A, the binding of purified Fab ANA15 to rabbit eEF1A purified from reticulocytes was examined. Rabbit eEF1A shares 100% aa homology with human eEF1A, but the two proteins exhibit differences in posttranscriptional methylation (27). As shown in Fig. 4, Fab ANA15 bound strongly to purified rabbit eEF1A. Strong binding was also observed with the rabbit anti-human eEF1A sera. In contrast, Fab ANA15 did not bind to irrelevant antigens, including ovalbumin, HIV-1 gp120, and RNA. Two anti-gp120 Fab fragments, L17 and b6, did not bind to the purified rabbit eEF1A.
Figure 4.
Binding of Fab ANA15 (5 μg/ml), donor serum (patient with Felty's syndrome, 1:200), polyclonal rabbit anti-human eEF1A-1 antibody (1:200), anti-gp120 Fabs L17 (negative control, 5 μg/ml) to purified intact rabbit eEF1A-1 by ELISA. To evaluate specificity, Fab ANA15 was also tested for binding to ovalbumin, HIV-1 gp120, and RNA.
We next examined colocalization between Fab ANA15 and the rabbit anti-human eEF1A serum on HEp-2 cells. As shown in Fig. 1 V–A′, some colocalization was observed. Both antibodies exhibited speckled staining of the nucleus, including the nucleoli. However, the rabbit antiserum exhibited weak cytoplasmic staining, whereas no staining of the cytoplasm was observed with Fab ANA15. The reason for this probably relates to the fact that RNA present in the nucleus influences the epitope of Fab ANA15. We have shown that the affinity of Fab ANA15 for eEF1A-1 is much higher when it is bound to RNA, whereas rabbit anti-human eEF1A sera contain antibodies that are both sensitive and insensitive to binding of eEF1A-1 to RNA. Detailed characterization of the specificity of Fab ANA15 will be described elsewhere (Ditzel et al., unpublished work).
Fab ANA15 Is Involved in an Active Immune Response.
Natural antibodies, mostly of the IgM class with low affinity and in germ line configuration, have been reported to bind self-antigens. To evaluate whether the IgG-derived Fab fragment ANA15 was derived from a plasma/B cell involved in an active immune response or was more akin to a natural antibody, the variable light and heavy chain genes were sequenced and compared with germ line sequences in the GenBank database. Both the variable light and heavy chain genes were somatically mutated, characteristic of an affinity-matured antibody. The variable light chain belongs to the VK3 subgroup and exhibits 87% nucleotide identity to kv325 as the closest germ line gene. The variable heavy chain belongs to the VH4 subgroup, exhibiting 94% nucleotide identity to the heavy chain germ line 71–4 as the closest germ line gene. Fab ANA15 uses the J segments JK2 and JH4b. Somatic mutations in the framework regions (FRs) and complementarity-determining regions (CDRs) were analyzed by measuring the replacement-to-silent mutation ratio for the CDR (CDR1 and CDR2) and FR (FR1, FR2, and FR3), which were 4.0 and 2.3 for the heavy chain and 3.75 and 1.0 for the light chain, respectively.
Autoantibodies involved in an active immune response are generally of higher affinity than natural antibodies. We therefore next determined the kinetic constants for the interaction of Fab ANA15 and recombinant human eEF1A-1 216–462 fragment by surface plasmon resonance. The values measured were kon = 5.0 × 104 M−1 s−1, koff = >10−6 s−1, resulting in a dissociation constant (Kd) of <0.02 nM.
To determine whether Fab ANA15 was a major contributor to the serum autoreactivity of the donor, the staining pattern of Fab ANA15 was then compared with that of the donor serum by using IF analysis and laser scanning confocal microscopy. Interestingly, Fab ANA15 and the donor serum showed distinctly different patterns of HEp-2 cell staining, demonstrating that ANA15 constituted only a minor part of the donor serum reactivity (Fig. 2). Six other antibody Fab fragments isolated from the Felty's syndrome antibody libraries were found, by comparative IF analysis, to correspond to the major response in the serum. Because biochemical analysis showed that these antibodies recognized annexin XI, a recently described autoantigen found in several autoimmune diseases (28), the use of cDNA library screening for antigen identification was not required. Detailed characterization of these antibodies will be described elsewhere. Similarly, the staining patterns of Fab ANA15 and the patient serum were compared on ethanol-fixed neutrophils. Both the Fab fragment and the serum stained the periphery of the nucleus of the neutrophils, and not lymphocytes (Fig. 2). Control sera from two SLE patients stained granulocytes and lymphocytes equally well, and sera from two healthy individuals showed no staining of any of the cells (data not shown). The serum of the donor from whom Fab ANA15 was cloned was also tested for binding to recombinant and purified eEF1A-1. At a dilution of 1:200, the donor sera exhibited strong staining of both antigens (Fig. 4).
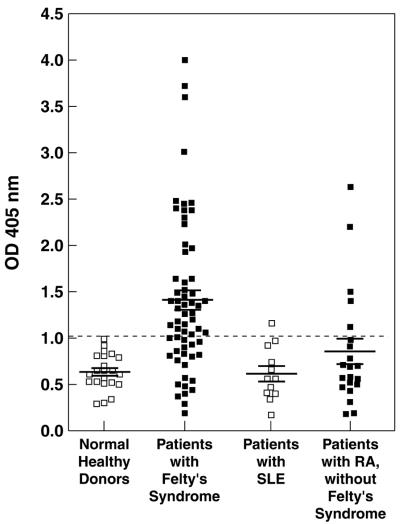
To investigate the frequency of patients with Felty's syndrome that had antibodies against eEF1A-1, a panel of 62 sera was titered for binding to purified eEF1A-1 in ELISA. Fig. 5 shows the binding data for sera diluted 1:100 from 62 patients with Felty's syndrome and 22 healthy normal donors. A strong correlation between Felty's syndrome and anti-eEF1A-1 IgG antibodies was observed as 41 of the 62 sera from Felty's syndrome patients and none of the sera of the healthy normal donors were considered positive. Positivity was defined as an OD405 value more than two standard deviations above the mean of the control normal donor values (>1.02). An alkaline phosphatase-conjugated F(ab′)2 fragment of a goat anti-human IgG Fc-specific antibody was used as secondary antibody to avoid problems of rheumatoid factor in the serum samples. In addition, 11 sera from patients with SLE and 21 sera from patients with RA without Felty's syndrome were studied. Only one sera from the group of SLE patients exhibited a marginally elevated level of anti-eEF1A-1 antibodies and 5 of the 21 sera (23%) from the patients with RA without Felty's syndrome exhibited anti-eEF1A-1 antibodies but with generally lower titers compared with the patients with Felty's syndrome. The mean level of anti-eEF1A-1 antibodies in the Felty's syndrome patients group was significantly different to the normal healthy donor group (P < 0.0001), whereas the difference between the mean anti-eEF1A-1 antibody level of the RA patients without Felty's syndrome and the SLE patient group was not significantly different to normal healthy individuals (P > 0.1 and P > 0.7, respectively). All of the sera were also tested against an irrelevant antigen, ovalbumin, to assess any contribution of polyreactive antibodies to reactivity with eEF1A-1. No significant difference between the percentage of sera with reactivity against ovalbumin from the different patient groups was observed. The numbers of sera that exhibited reactivity with ovalbumin were: Felty's syndrome, 16/62; healthy donors, 8/22; SLE, 3/11; and RA without Felty's syndrome, 4/21. The lack of correlation between reactivity with eEF1A-1 and ovalbumin, indicated that polyreactive behavior could not explain serum reactivity with eEF1A-1 in the majority of Felty's syndrome patients. The 62 Felty's syndrome sera diluted 1:20 were also tested for ANA staining. Forty of the 62 patients (65%) had positive ANA. There was a good correlation between patients having positive ANA and eEF1A-1 antibodies, as 38 patients were positive in both assays, whereas 2 exhibited positive ANA but negative anti-eEF1A-1 antibodies and 4 had detectable anti-eEF1A-1 antibodies but were negative in the ANA test.
Figure 5.
Serum antibody reactivity (1:100) against eEF1A-1 as measured by ELISA for 62 patients with Felty's syndrome, 21 patients with RA without Felty's syndrome, 11 patients with SLE, and 22 normal healthy donors. The mean and standard error for each group are shown as long and short horizontal lines, respectively.
Discussion
Valuable information concerning the mechanism(s) leading to autoimmune disease has been generated by using animal models (3, 6, 29). However, in other instances, the relevance to human disease of results obtained in autoimmune animal models has been questioned. Studies in the human system, on the other hand, are more difficult, but the information obtained is more clinically relevant. In humans, a major obstacle to progress is the identification and cloning of the autoantibodies and autoantigens that may be critical for disease development and progression. To identify and clone these molecules, we developed an approach whereby an antibody phage display library reflecting the serum response of the donor is generated and selected against target cells such as diseased tissue. Cloned phage antibodies are then used as probes for the isolation of cDNA clones that encode the corresponding autoantigen. We used this approach to study the autoantibody–antigen repertoire in a patient with Felty's syndrome, an extraarticular manifestation of RA with peripheral destruction of neutrophils. The success of this approach lead us to identify and clone eEF1A-1–anti-eEF1A-1 as a novel autoantigen–antibody pair in this disease. Subsequently, the relevance of eEF1A-1 in Felty's syndrome was demonstrated by the finding that 66% of patients with Felty's syndrome exhibit anti-eEF1A-1 antibodies. In contrast, only 1 of 11 patients with SLE exhibited marginally increased anti-eEF1A-1 titers. Furthermore, 5 of 21 patients with RA without Felty's syndrome exhibit anti-eEF1A-1 antibodies but generally at lower titers than observed with the Felty's syndrome patients.
Other approaches for the identification of novel human autoantigens have been described, including the use of patient sera to probe recombinant expression libraries (SEREX). This approach has been used to clone several very interesting antigens, including cancer-associated antigens in particular (11). However, the full spectrum of autoantigens may not be identified in any given case by SEREX because of the low concentrations of some autoantibodies and/or interfering proteins in the whole serum (12). The use of antibody phage display in the initial step may allow a more detailed analysis of the autoantibody–antigen repertoire in a given donor. This approach does involve extra effort when compared with the SEREX approach in the form of antibody library construction and selection. However, the cloning of the serum antibodies should allow for easier screening of the cDNA expression libraries by avoiding the two major complicating factors associated with SEREX: the presence of naturally occurring antibacterial antibodies in serum and the presence of cDNAs in the tissue-derived expression libraries that encode for endogenous IgG (12). Both of these factors can result in false-positive cDNA clones. The false-positive IgG clones can be avoided in the approach adapted here by tagging the cloned antibody fragment and detecting its binding with an antitag antibody to eliminate the use of anti-IgG antibodies in library screening. Furthermore, because the antibody phage display system selects for antibody–antigen interactions of higher affinity when compared with SEREX, where high levels of antibodies of moderate affinity may dominate the selection process, more disease-relevant antibody–antigen pairs may be identified by this new approach. This seems to be the case for the anti-eEF1A-1 antibody derived from the patients studied here. Comparison of the binding pattern of the cloned antibody with the polyclonal serum of the patient demonstrated that anti-eEF1A-1 represented only a minor part of the serum autoimmune response.
The identification of eEF1A-1 as a novel target for ANA and antineutrophil antibodies and its strong association with Felty's syndrome may have important clinical relevance. Previous observations have suggested that ANA antibodies of yet undefined specificities are prevalent in the sera of 70% of patients with Felty's syndrome in contrast to 30% of RA patients without Felty's syndrome (20, 22). Our observation that 41 of 62 (66%) of Felty's syndrome patients exhibited serum anti-eEF1A-1 antibodies and the existence of a strong correlation between anti-eEF1A-1 antibodies and positive ANAs are consistent with the notion that these antibodies correspond to a major part of the unidentified ANA fraction.
The question remains why antibodies to eEF1A-1 are generated in a high percentage of patients with Felty's syndrome. Because Fab ANA15 is derived from an IgG library, is of high affinity and shows evidence of extensive somatic mutation with high replacement-to-silent mutation ratio in the CDR regions, it appears that the antibody is produced as a result of a matured immune response to eEF1A-1. Furthermore, the ANA15 antibody was probably produced by actively secreting plasma cells, because the IgG antibody phage display library used for this study was constructed by using RNA isolated from bone marrow cells of a Felty's syndrome patient. While investigating the specificity of ANA15 antibody, we interestingly found that the affinity of ANA15 for purified rabbit eEF1A was lower than that for the E. coli-expressed eEF1A-1 fragment. The difference probably reflects the fact that the latter protein was complexed to RNA; RNA has been found to strongly enhance the binding of ANA15 to eEF1A-1 (Ditzel et al., unpublished observations). The involvement of RNA in effective Fab ANA15 binding to eEF1A-1 is also the likely reason for Fab ANA15 staining of the nucleus and not the cytoplasm, the cell compartment where the translation process takes place. The presence of eEF1A-1 in the nucleus has been suggested by others (30). Furthermore, the polyclonal serum raised by immunization of rabbits with eEF1A-1 reacted both with the cytoplasm and nucleus. At least two possible mechanisms may explain why antibodies are generated against an antigen such as eEF1A-1 which under normal condition is located intracellularly. One explanation is that cells in the disease process express eEF1A-1 on the cell surface. It has, for example, been shown that during apoptosis of neutrophils, intracellularly located myeloperoxidase is translocated to the cell surface where it is exposed to the antimyeloperoxidase-type antineutrophil cytoplasmic antibodies antibodies (10). We examined this scenario by induction of apoptosis of purified neutrophils by extended in vitro culture or irradiation with UV light. In apoptotic neutrophils, staining with Fab ANA15 was found at the cell surface and just beneath the cell surface. The strongest staining was observed in surface blebs of the apoptotic cells (Ditzel et al., unpublished work). The translocation of eEF1A-1 from the nucleus to the cell surface of apoptotic cells is in agreement with observations on the translocation of other autoantigens in apoptotic cells (9, 10).
In conclusion, we have identified and cloned eEF1A-1 and a human anti-eEF1A-1 antibody as a novel autoantigen–antibody pair and demonstrated a strong association between anti-eEF1A-1 antibodies and Felty's syndrome. These results were obtained by using an approach based on the initial dissection of the immune response by phage display technology and subsequent cloning of the corresponding antigen from cDNA expression libraries. The approach should be generally applicable and allow identification of both cell surface, cytoplasmic, and as in this case, nuclear antigens. Cloning and expression of human autoantigens and antibodies should permit systematic studies of any pathogenicity resulting from the antigen–antibody interaction.
During the revision of this manuscript, a report was published on the presence of anti-eEF1A antibodies in a small percentage of patients with atopic dermatitis (31). Interestingly, the authors found, in agreement with our observations, that patients with anti-eEF1A antibodies had low white blood cell counts. However, no detailed analysis of the type of white blood cell that was affected was performed. We speculate that the observed anti-eEF1A antibodies in the study of Ohkouchi et al. (31) relate to neutrophil destruction and lowered white blood cell counts in the patients described rather than atopic dermatitis.
Acknowledgments
We thank Robert Fox, Paul Parren, and Pascal Poignard for helpful discussions. This work was supported by National Institutes of Health Grant AI041590 to H.J.D.
Abbreviations
- eEF1A-1
eukaryotic elongation factor 1A-1
- SLE
systemic lupus erythematosus
- RA
rheumatoid arthritis
- SEREX
serum analysis of recombinant expression libraries
- ANA
antinuclear antigen
- FR
framework region
- CDR
complementarity-determining region
- IF
immunofluorescence
Footnotes
References
- 1.Tan E M, Cohen A S, Fries J F, Masi A T, McShane D J, Rothfield N F, Schaller J G, Talal N, Winchester R J. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 2.Greaves M. Lancet. 1999;353:1348–1353. doi: 10.1016/S0140-6736(98)10362-8. [DOI] [PubMed] [Google Scholar]
- 3.Ziporen L, Shoenfeld Y, Levy Y, Korezyn A D. J Clin Invest. 1997;100:613–619. doi: 10.1172/JCI119572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierangeli S S, Colden-Stanfield M, Liu X, Barker J H, Anderson G L, Harris E N. Circulation. 1999;99:1997–2002. doi: 10.1161/01.cir.99.15.1997. [DOI] [PubMed] [Google Scholar]
- 5.Funauchi M, Ikoma S, Enomoto H, Ohno M, Kinoshita K, Horiuchi A, Kurata N. Intern Med. 1996;35:367–372. doi: 10.2169/internalmedicine.35.367. [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Nagy Z, Radic M Z, Hardy R R, Huszar D, Camper S A, Weigert M. Nature (London) 1995;373:252–255. doi: 10.1038/373252a0. [DOI] [PubMed] [Google Scholar]
- 7.Korganow A-S, Ji H, Mangialaio S, Duchatelle V, Pelanda R, Martin T, Degott C, Kikutani H, Rajewsky K, Pasquali J L, et al. Immunity. 1999;10:451–461. doi: 10.1016/s1074-7613(00)80045-x. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto I, Staub A, Benoist C, Mathis D. Science. 1999;286:1732–1735. doi: 10.1126/science.286.5445.1732. [DOI] [PubMed] [Google Scholar]
- 9.Casciola-Rosen L A, Anhalt G, Rosen A. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilligan H M, Bredy B, Brady H R, Hebert M J, Slayter H S, Xu Y, Rauch J, Shia M A, Koh J S, Levine J S, et al. J Exp Med. 1996;184:2231–2241. doi: 10.1084/jem.184.6.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M, et al. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Old L J, Chen Y T. J Exp Med. 1998;187:1163–1167. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folgori A, Tafi R, Meola A, Felici F, Galfré G, Cortese R, Monaci P, Nicosia A. EMBO J. 1994;13:2236–2243. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williamson R A, Burioni R, Sanna P P, Partridge L J, Barbas C F, Burton D R. Proc Natl Acad Sci USA. 1993;90:4141–4145. doi: 10.1073/pnas.90.9.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton D R, Barbas C F, Persson M A A, Koenig S, Chanock R M, Lerner R A. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ditzel H J, Binley J M, Moore J P, Sodroski J, Sullivan N, Sawyer L S W, Hendry R M, Yang W-P, Barbas C F, Burton D R, et al. J Immunol. 1995;154:893–906. [PubMed] [Google Scholar]
- 17.De Kruif J, Terstappen L, Boel E, Logtenberg T. Proc Natl Acad Sci USA. 1995;92:3938–3942. doi: 10.1073/pnas.92.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marks J D, Ouwehand W H, Bye J M, Finnern R, Gorick B D, Voak V, Thorpe S J, Hughes-Jones N C, Winter G. Bio/Technology. 1993;11:1145–1148. doi: 10.1038/nbt1093-1145. [DOI] [PubMed] [Google Scholar]
- 19.Starkebaum G, Arend W P, Nardella F A, Gavin S. J Lab Clin Med. 1980;96:238–251. [PubMed] [Google Scholar]
- 20.Coremans I E, Hagen E C, van der Voort E A, van der Woude F J, Daha M R, Breedveld F C. Clin Exp Rheumatol. 1993;11:255–262. [PubMed] [Google Scholar]
- 21.Rothko K, Kickler T S, Clay M E, Johnson R J, Stroncek D F. Blood. 1989;74:1698–1703. [PubMed] [Google Scholar]
- 22.Juby A, Johnston C, Davis P, Russell A S. Br J Rheumatol. 1992;31:185–188. doi: 10.1093/rheumatology/31.3.185. [DOI] [PubMed] [Google Scholar]
- 23.Starkebaum G, Singer W P, Arend W P. Clin Exp Immunol. 1980;39:307–314. [PMC free article] [PubMed] [Google Scholar]
- 24.Breedveld F C, Lafeber G J M, De Vries E, Van Krieken J H J M, Cats A. Ann Rheum Dis. 1986;45:696–702. doi: 10.1136/ard.45.8.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbas C F I, Kang A S, Lerner R A, Benkovic S J. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlsson R A, Michaelsson A, Mattsson L. J Immunol Methods. 1991;145:229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 27.Merrick W C, Dever T E, Kinzy T G, Conroy S C, Cavallius J, Owens C L. Biochim Biophys Acta. 1990;1050:235–240. doi: 10.1016/0167-4781(90)90173-y. [DOI] [PubMed] [Google Scholar]
- 28.Misaki Y, Pruijn G J M, van der Kemp A W C M, van Venrooij W J. J Biol Chem. 1994;269:4240–4246. [PubMed] [Google Scholar]
- 29.Burkhardt H, Kalden J R. Rheumatol Int. 1997;17:91–99. doi: 10.1007/s002960050015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders J, Maassen J A, Möller W. Nucleic Acids Res. 1992;20:5907–5910. doi: 10.1093/nar/20.22.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohkouchi K, Mizutani H, Tanaka M, Takahashi M, Nakashima K, Shimizu M. Int Immunol. 1999;11:1635–1640. doi: 10.1093/intimm/11.10.1635. [DOI] [PubMed] [Google Scholar]