Abstract
Type 1 pilus biogenesis was used as a paradigm to investigate ordered macromolecular assembly at the outer cell membrane. The ability of Gram-negative bacteria to secrete proteins across their outer membrane and to assemble adhesive macromolecular structures on their surface is a defining event in pathogenesis. We elucidated genetic, biochemical, and biophysical requirements for assembly of functional type 1 pili. We discovered that the minor pilus protein FimG plays a critical role in nucleating the formation of the adhesive tip fibrillum. Genetic methods were used to trap pilus subunits during their translocation through the outer membrane usher protein, providing data on the structural interactions that occur between subunit components during type 1 pilus formation. Electron microscopic and biochemical analyses of these stepwise assembly intermediates demonstrated that translocation of pilus subunits occurs linearly through the usher's central channel, with formation of the pilus helix occurring extracellularly. Specialized pilin subunits play unique roles both in this multimerization and in the final ultrastructure of the adhesive pilus.
Type 1 pili, one of more than 30 different adhesive structures built by chaperone/usher-dependent pathways, contain the FimH adhesin, which is critical in the ability of the uropathogenic Escherichia coli to cause cystitis (bladder infections; refs. 1–3). FimH mediates colonization by binding to mannose-containing uroplakin receptors on the luminal surface of bladder epithelium (3). Superficial bladder cells exfoliate in response to E. coli infection, which has been proposed to be an important host defense mechanism to prevent the infection from establishing a foothold (3). However, FimH-mediated colonization and invasion of the exposed underlying tissue is one mechanism by which uropathogenic E. coli subvert the exfoliation response and cause a persistent infection (3).
In addition to the chaperone/usher pathway, many plant and animal pathogens such as Salmonella, E. coli, and Yersinia employ other secretion systems, including the type III secretion pathway. The type III secretion machinery is used for the secretion and translocation of numerous different cytotoxins into eukaryotic cells (reviewed in refs. 4 and 5). In addition, type II secretion, or the general secretion pathway, is used to secrete toxins and construct type IV pili in a variety of pathogenic organisms including Neisseria, Klebsiella, and Pseudomonas (reviewed in refs. 6 and 7). There are clearly many differences between each of the secretion systems, but similarities have begun to emerge. Interestingly, type II, type III, and chaperone/usher-dependent secretion systems all result in the formation of macromolecular structures (7–12). These systems, as well as the filamentous phage assembly pathway, use ultrastructurally similar oligomeric, pore-forming, possibly gated, outer membrane proteins to facilitate outer membrane translocation (13–18).
Extensive research into protein secretion systems has primarily centered around events before translocation across the outer membrane and on the ultimate pathogenic role of the secreted proteins. For example, in E. coli, chaperone/usher-dependent type 1 pili are encoded by an operon consisting of eight genes, FimA–FimH (ref. 19; Fig. 1). Type 1 pili are composite structures consisting of short tip fibrillae joined to the distal ends of pilus rods (10). The pilus rod is a helical, 70-Å-wide cylindrical structure comprised primarily of FimA subunits. The tip fibrillum is comprised of the mannose-binding FimH adhesin and the minor pilins FimG and FimF. FimF and FimG seem to play some role in the regulation of length and number of pili (10, 20, 21), but their functions and any mechanisms of action remain equivocal. The crystal structure of the FimC periplasmic chaperone in a complex with the FimH adhesin revealed that FimC is comprised of two Ig-like domains oriented in an L shape (22). FimH also consists of two domains, an Ig-like pilin domain and a receptor-binding domain that has a jelly roll-like fold (22). The pilin domain of FimH has a similar structure to the PapK pilin of the P pilus system; both lack the C-terminal G β-strand, creating a deep groove on the surface of the folded pilin (23). Pilins require a chaperone to provide this steric information, in the form of the G1 β-strand, to facilitate proper folding in a process called donor strand complementation (22–24). FimC remains bound to the folded protein, with its G1 β-strand occupying the groove of the folded protein, thus preventing nonproductive aggregation in the periplasm (22, 23). Periplasmic chaperone-subunit complexes are targeted to FimD, an outer membrane molecular usher, which facilitates chaperone uncapping and the translocation of pilus subunits across the outer membrane and their assembly into pili (17, 25). In the mature pilus, a conserved N-terminal extension of each subunit is thought to complete the fold of its neighbor more permanently by occupying the groove in a process called donor strand exchange (22–24), a process that depends on the outer membrane usher.
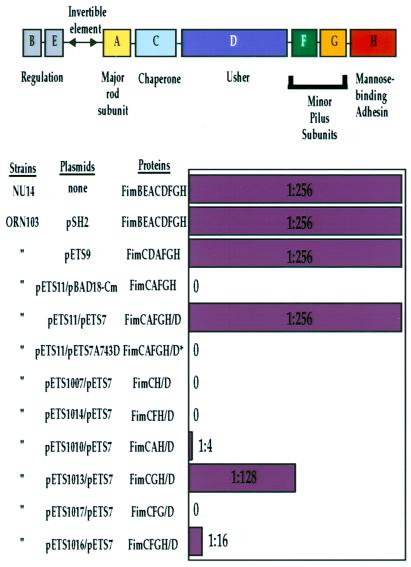
Figure 1.
Genes necessary for functional pili. The type 1 pilus gene cluster and gene products necessary for the expression and assembly of type 1 pili are shown (reviewed in ref. 38). See Introduction for a description of the gene products represented as A–H in the schematic depiction of the fim operon shown at the top. The bar graph indicates the ability of various combinations of type 1 genes to encode for production of functional type 1 pili as assessed by the hemagglutination (HA) of guinea pig red blood cells (39). The HA titer produced by ORN103/pETS9 (1:256) was identical to that of a type 1 operon under the control of its wild-type promoter or an NU14 clinical isolate. In a two-plasmid system, the HA titer matched that of the wild type, whereas with a nonfunctional mutant fimD (pETS7A743D) no HA occurred (see Materials and Methods for information regarding mutant).
The data from the following studies provide insight into vital and undefined events in type 1 pilus biogenesis: translocation of pilins across the outer membrane and their assembly into pili. Through examination of macromolecular assembly products copurified with the outer membrane usher protein FimD, we demonstrate that the subunits of the chaperone/usher-dependent type 1 pili system traverse the outer membrane in a linear fashion through the pore of the usher. The pilus seems to coil into its helical form only after exiting the pore. The usher-based assembly intermediates have also helped us define specific functions of individual protein components as they cross the outer membrane and oligomerize to form a macromolecular assembly. Although the pilins that comprise type 1 pili are homologous, it is their differences that define the specific role of each within the final macromolecular assembly. The characterization of these assembly intermediates has provided insight into the molecular basis of the regulation of usher-mediated protein translocation and ordered pilus assembly, as well as providing further understanding of a pathogenic cascade that may exploit the type 1 pilus architecture.
Materials and Methods
Strains and Plasmids.
The NU14 clinical isolate and the type 1− strain ORN103 have been described (2, 19, 26). The plasmid pETS7 was made by subcloning histidine-tagged fimD from pETS4 (17) into the arabinose-inducible plasmid pBAD18-Cm (27). All of the vectors containing chaperone–subunit genes were made by subcloning respective genes from pSH2 and pHJ20 (10, 19) into the isopropyl β-d-thiogalactoside-inducible plasmid pMMB66 (28). Orientation of genes was checked by restriction digest and protein expression. The plasmid pETS7A743D contains a mutant fimD with one amino acid change (A743D) and is completely unable to assemble pili (unpublished data).
Protein Production and Purification.
Pilus production from NU14 and pSH2/ORN103 was performed as described (2, 10, 26). Protein production/cell fractionation/protein solubilization/gel filtration was done as described for FimD usher alone and the FimDCH complex (17). Cells and proteins were never frozen before electron microscope studies. In a negative control experiment, we were unable to copurify any intermediates with the A743D mutant FimD (data not shown).
Electron Microscopy.
For negatively stained samples (1% uranyl acetate), fractions from gel filtration were diluted 2:400 in 20 mM Tris (pH 8.0)/75 mM NaCl. For unidirectional shadowing, the sample was in seven parts glycerol and three parts 1 M ammonium acetate and was shadowed at an angle of 11–15° with evaporated platinum. After digitization of negatives, color tables were adjusted such that the slope of the curves was ≥0, with zero or one points of inflection. The images shown were chosen to be good representations of the thousands of particles observed in the course of these studies.
Miscellaneous.
HA assays and Western blots were done as described (10, 17, 26). The anti-FimH antibodies (raised against an N-terminal 175-amino acid FimH truncate) and the anti-type 1 pilus antibodies were kindly provided by Medimmune (Gaithersburg, MD). Anti-type 1 pilus antibodies do not recognize FimC or FimD (data not shown). Soman Abraham's lab (Duke Univ., Durham, NC) kindly provided the anti-FimG antibodies (raised against a peptide containing the N-terminal 21 amino acids of FimG).
Results
Subunits Required for FimH-Mediated HA.
The initial step of pilus assembly seems to involve the kinetic partitioning of FimCH chaperone–adhesin complexes to a periplasmic region of the FimD usher (17). Formation of the FimDCH ternary complex is necessary to initiate pilus assembly (17), but cells expressing only FimDCH are not able to hemagglutinate guinea pig erythrocytes (Fig. 1). FimA, FimG, and FimF were coexpressed with FimDCH in a variety of combinations to test the minimum requirements necessary to trigger the translocation of FimH across the membrane in a manner that makes it accessible to bind mannose, as measured by HA of guinea pig erythrocytes. Coexpression of FimG (but not FimF or FimA) with FimDCH resulted in an almost wild-type HA titer (Fig. 1). A sequential series of assembly intermediates was purified and characterized to investigate the protein–protein interactions necessary for translocation and multimerization of the pilins through the FimD usher. These intermediates essentially represent snapshots of virulence protein translocation across the outer membrane of bacteria. Fig. 2 represents a biochemical analysis, and Fig. 3 illustrates an electron microscopic analysis of these intermediates. In the text, the description of biochemical intermediates in Fig. 2 is coupled with a corresponding discussion of the electron microscopic image of that intermediate, shown in Fig. 3, to reflect the ordered events in pilus biogenesis.
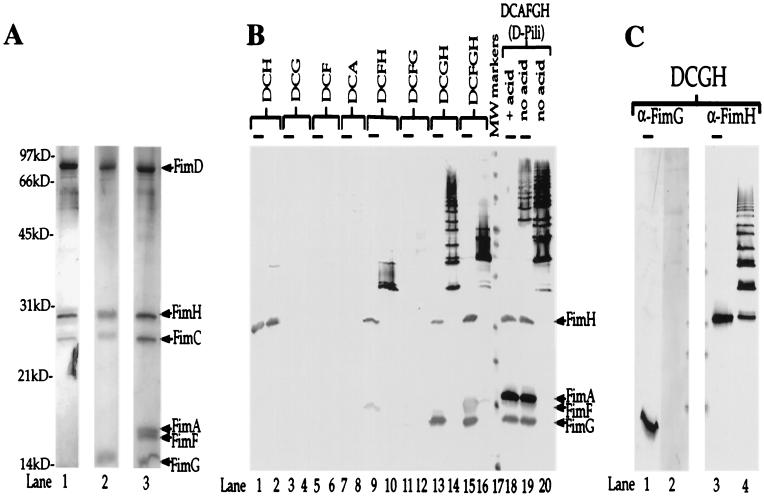
Figure 2.
Multimerization events demonstrate molecular specificity of subunit-subunit interactions. (A) Silver-stained gel (Bio-Rad Silver Stain Plus). Outer membrane preparations were subjected to nickel chromatography (to purify the His-tagged FimD) after coexpression with various combinations of pilus subunits. Samples were boiled in SDS sample buffer and subjected to standard SDS/PAGE. Protein samples purified from E. coli expressing only FimDCH were run in lane 1; those expressing only FimDCGH were run in lane 2; and those expressing FimDCAFGH were run in lane 3. Arrows and labels indicate the expected molecular masses of each of the copurifying proteins: FimD, 92 kDa; FimH, 29 kDa; FimC, 26 kDa; FimA, 18 kDa; FimF, 17 kDa; and FimG, 15 kDa. kD, kilodalton. (B) Anti-type 1 pilus Western blot of samples (purified as those shown in A) either boiled (lanes with overlying bars: odd numbered lanes 1–15 and lanes 18 and 19) or kept at room temperature (even numbered lanes 2–20 with the exception of lane 18) in SDS sample buffer before standard SDS/PAGE. Based on the behavior of whole pili in this assay (lanes 18–20) and previous studies (10), higher-molecular-mass ladders represent higher-order multimers. (C) Western blot of the same samples analyzed in B, lanes 13 and 14, except the blot was developed with anti-FimG antiserum (prepared against an N-terminal 21-amino acid peptide; lanes 1 and 2) and anti-FimH antiserum (lanes 3 and 4). Lanes 2 and 4 contain unboiled samples. Lanes 1 and 3 contain boiled samples (lanes with overlying bars). We hypothesize that the N terminus of FimG is hidden in the FimH-FimG multimer, because it is likely involved in head-to-tail donor strand exchange interactions (22–24, 31).
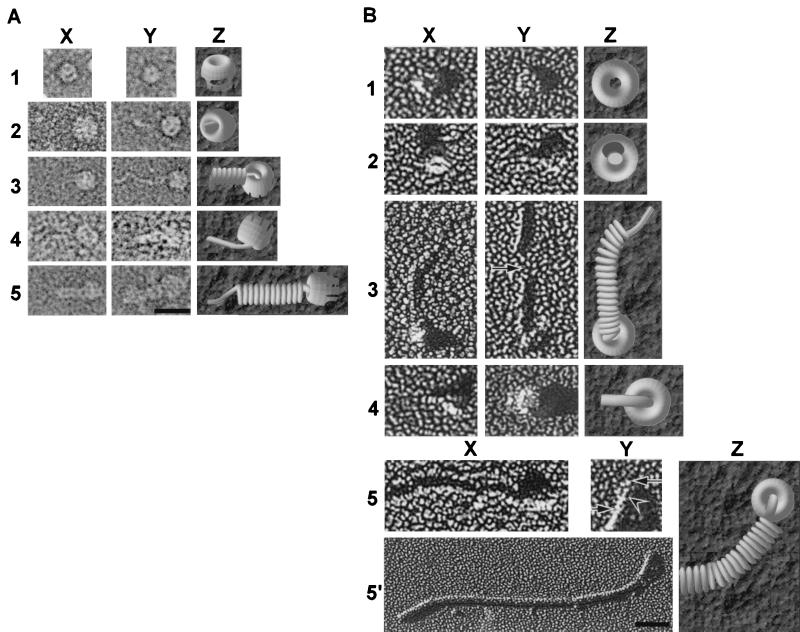
Figure 3.
Electron microscopic images of usher-based assembly intermediates. (A and B) Row 1, FimD alone; row 2, FimDCH complexes; row 3, FimDCGH complexes; row 4, FimDCFGH complexes; row 5 (and 5′ in B), FimD–pili (FimDCAFGH) complexes. Column Z contains models of the various assembly intermediates shown in column X, except the model in 5Z, which is a model of 5Y and 5′. (A) Negatively stained complexes. The images shown in row 1 illustrate two orientations of the usher. Face-on views (1Y) are common only in samples with FimD alone. The usher is positioned at the right-hand side of the images in rows 2–5 with fibers extending to the left. (B) Unidirectionally shadowed complexes. The ushers are positioned as in A except in 2X, 4X, and 4Y, in which the fibers are extending out toward the reader. The ushers in row 3 are at the bottom, with fibers extending upward. Most ushers adhered to the grid en face; however, an edge-on view is seen in 3Y. The arrow in 3Y indicates a portion of unraveled helical rod structure in the FimDCGH assembly intermediate. 5Y is an enlarged view of the usher seen in row 5′. The long arrow indicates the ring-shaped usher; the short arrow indicates the pilus rod; the arrowhead indicates a linear fiber emerging from the central pore of the usher. (Bar in A = 250 Å, applicable to all of the images except 5′; bar in 5′ = 500 Å.)
FimDCH Usher–Chaperone–Adhesin Ternary Complex.
As was shown previously (17), FimCH chaperone–adhesin complexes (Fig. 2A, lane 1; Fig. 2B, lanes 1 and 2) but not FimCA, FimCF, or FimCG complexes (Fig. 2B, lanes 3–8) could be copurified with the FimD usher. A small amount of FimH appeared as a stable, higher-molecular-mass complex when the FimDCH sample was incubated in SDS at room temperature but was dissociated on boiling in SDS (compare Fig. 2B, lane 2, with Fig. 2 A and B, lane 1). When FimH is not present, as in a strain expressing FimDCFG, neither FimG nor FimF can be copurified with the FimD usher (Fig. 2B, lanes 11 and 12). This finding further supports the theory that the preformed FimDCH usher–chaperone–adhesin complex is required to prime the usher for pilus assembly and stabilizes the usher in its active form (17). With electron microscopy, we saw that the purified FimD usher exists in cylindrical, ring-shaped structures that are approximately the same size and shape as the P pilus usher PapC (29), with a 150-Å outer diameter and a 20-Å central pore (Fig. 3 A and B, row 1). It seems that one end of the cylindrical usher forms a scaffold-like structure (e.g., Fig. 3A 1X, 2Y, and 5X), whereas the other end is a more electron-dense structure (Fig. 3A 1X and 5X). The purified FimDCH usher–chaperone–adhesin complexes appear as cylindrical objects indistinguishable from the FimD samples (for example, Fig. 3A, compare 1X and 2Y, or Fig. 3B, compare 1X and 2X), except that there were often nubs filling the central pore [Fig. 3A 2X (model in 2Z); Fig. 3B 2X (model in 2Z)]. These data strongly suggest that pilus protein translocation occurs through the central pore of the usher—a mechanism that has been proposed previously but never shown (e.g., refs. 18 and 29). In the FimDCH sample, short, fibrillar tip-like structures 20 Å in diameter (Fig. 3A 2Y; Fig. 3B 2Y) were observed occasionally. Such a thin, fibrillar structure could be produced by two or three FimH molecules based on the dimensions of FimH revealed in the crystal structure, which was 20 Å wide × 110 Å long (22). The biochemical analysis of the FimDCH sample also revealed that a small percentage of the material ran as a multimeric species, most likely a dimer (Fig. 2B, lane 2). The presence of FimG and the other pilus subunits in a wild-type pilus may preclude such FimH–FimH interactions in favor of potentially more stable and complementary subunit–subunit interactions.
Multimerization Triggered by FimG.
When FimDCGH proteins were expressed, FimG copurified with FimD along with FimH and FimC (Fig. 2A, lane 2), and a high-order multimer stable in SDS at room temperature was formed (Fig. 2B, lane 14). The monomeric FimG and FimH bands present in the boiled sample (Fig. 2B, lane 13) both disappear in the room temperature sample (Fig. 2B, lane 14), suggesting that both FimG and FimH are present in the oligomeric ladder. The presence of FimH in the ladder was confirmed by Western blotting (Fig. 2C, lanes 3 and 4). A Western blot with an antibody directed against the N-terminal 21 amino acids of FimG was not able to recognize FimG in the ladder (Fig. 2C, lanes 1 and 2). This result is consistent with previous results (30), and the hypothesis that the extreme N terminus of subunits is “hidden” in the multimerized structure because of “head-to-tail” interactions between subunits (22–24, 31).
Electron microscopic analysis of the FimDCGH complexes revealed that they form diverse structures despite the minimal number of proteins (Fig. 3 A and B, row 3). We frequently observed complexes that looked somewhat like cherries—with a cylindrical protein complex at the base of a linear structure (Fig. 3A 3Y). These complexes likely represent an approximately 20-Å diameter FimG–FimH structure growing through the central pore of an oligomeric, cylindrical usher. In addition, we consistently saw complexes in which the tip structure extending from the cylindrical usher appeared to wrap into a pilus-like rod having an ≈70-Å diameter [Fig. 3A 3X (model in 3Z); Fig. 3B 3X and 3Y]. These rod-like forms often contained kinks [Fig. 3B 3X (model in 3Z)] or even unwound regions in the middle of the rod (Fig. 3B 3Y, arrow). The different morphologies of the rod form suggest that there may be inherent structural heterogeneity.
Termination of Multimerization by FimF.
FimG–FimH multimer export and assembly seem to be terminated by the addition of FimF, as evidenced by the decrease in HA titer (Fig. 1) and the decrease in higher-order oligomers when FimF is expressed (Fig. 2). FimF did copurify when expressed with FimH as a FimDCFH complex (Fig. 2B, lanes 9 and 10), but it did not copurify with the FimD usher when expressed from fimDCF (Fig. 2B, lanes 5 and 6). The FimF and FimH bands seen in the sample boiled in SDS (Fig. 2B, lane 9) existed as somewhat higher-molecular-mass complexes at room temperature but significantly higher-order oligomers were absent (Fig. 2B, lane 10). Thus, FimF, like FimG, is capable of interacting with the FimDCH ternary complex. However, unlike the FimDCGH interaction, further multimerization events do not occur efficiently. When FimF was expressed along with FimDCGH (as FimDCFGH), the size of the higher-order oligomers decreased compared with FimDCGH alone (Fig. 2B, compare lanes 14 and 16), suggesting that FimF terminates FimGH oligomerization. As determined by electron microscopy, FimDCFGH complexes (Fig. 3 A and B, row 4) looked much like the FimDCGH complexes—cylindrical protein complexes attached to a thin fibrillum [Fig. 3A 4X (model in 4Z); Fig. 3B 4X (model in 4Z)] that sometimes were wrapped into thicker, rod-like fibers (Fig. 3A 4Y; Fig. 3B 4Y).
Characterization of Usher–Pilus Fiber Complexes.
When FimDCAFGHs (whole pili) were expressed, FimA, FimF, FimG, FimH, and FimC all copurified with the FimD usher (Fig. 2A, lane 3; Fig. 2B, lanes 18–20). The dissociation of type 1 pili requires their boiling in SDS sample buffer containing concentrated hydrochloric acid (19). Strong FimA–FimA quaternary interactions make the pilus rods extremely stable, with every subunit donating its N-terminal extension to complete the Ig fold of its neighboring subunit (22, 23). In the absence of acid, the purified material from the FimDCAFGH (whole pili) strain showed a high-molecular-mass ladder (Fig. 2B, lanes 19 and 20) when analyzed by SDS/PAGE and immunoblotting with anti-type 1 pilus antisera. This ladder, indicative of FimA oligomers in a pilus rod (32), was dissociated only partially by boiling, and full dissociation into monomers required boiling in acid (Fig. 2B, lane 18). The FimC chaperone copurified with every assembly intermediate but did not copurify with the FimD usher from a strain expressing only FimC and FimD (17). These data further our hypothesis that the chaperone remains capped to the last subunit incorporated into the growing pilus, with its G1 β-strand presumably occupying the groove of the subunit. The chaperone's participation in donor strand complementation (22, 23) precludes the premature uncapping of the chaperone, because uncapping would expose the hydrophobic core of the pilin, leading to its denaturation. In our expression system, most FimD–pilus complexes appeared as short pili attached to the usher (Fig. 3 A and B, row 5 and 5′). Sometimes, the pili clearly appeared in a linear form closest to the usher and a rod form further away (Fig. 3B 5Y, which is an enlarged region of the usher from the image in 5′ and is modeled in 5Z). Such images provide direct evidence for the theory (29, 33) that rod-forming pili are sterically forced to grow through the multimeric usher in a linear fashion and thereafter snap into their characteristic right-handed helical conformation. Thus, a quaternary rearrangement must occur in the pilus fiber to produce a 70-Å-diameter filament after the pilin subunits transit through a 20-Å-diameter usher pore.
Discussion
The type 1 pilus system is one of many protein secretion/macromolecular assembly systems used by Gram-negative bacteria to facilitate infection of a host. Previous studies have alluded to the possibility that protein translocation across the outer membrane of Gram-negative bacteria occurs through the pore of the required outer membrane protein. By obtaining freeze frames of pilus assembly, we have demonstrated that the ubiquitous process of protein translocation occurs through the central pore of the oligomeric usher protein in the type 1 pilus system. In the type 1 system—and likely in each of the more than 30 different chaperone/usher-dependent secretion systems—translocation and assembly occur in a linear fashion, and the adoption of a helical pilus rod form occurs subsequent to the translocation event.
The data described above suggest that the interaction of FimG with the FimDCH ternary complex nucleates the formation of a FimH-containing tip fibrillum and/or rod structure that is translocated across the outer membrane in an usher-dependent manner. Based on the recently solved crystal structure of the FimCH chaperone–adhesin complex (22), the lectin (sugar-binding) domain of FimH is presumably inserted into the usher channel with the pilin domain remaining in the periplasm bound via donor strand exchange to the chaperone, which completes its Ig fold. The groove in FimH pilin domain has been proposed to represent the interactive tail of the subunit that then participates in a head-to-tail interaction with a neighboring subunit in the pilus after chaperone uncapping (22–24, 31). The pilin head has been proposed to be comprised of a conserved N-terminal extension present in all pilus subunits (22, 23, 31). For example, FimG from an incoming FimCG complex may replace the G1 β-strand of the chaperone with its N-terminal extension (22–24), thereby displacing the chaperone and driving pilus assembly. Formation of specific, energetically favorable subunit–subunit interactions could nucleate multimerization of the FimG–FimH tip fibrillum through the central pore of the FimD usher. The role of the scaffold-like periplasmic or transmembrane regions of the FimD usher in guiding assembly remains to be determined.
The FimC–FimH crystal structure shows that the pilin domain of FimH possesses a groove because of the lack of a C-terminal G1 β-strand (22). Although FimH is lacking an N-terminal extension, it instead possesses a receptor-binding domain linked to the pilin domain by a short 4-amino acid peptide sequence (22). The presence of a pilin groove and the lack of an N-terminal extension suggest a mechanism by which a single FimH is located only at the tip of the pilus. Tip fibrillae have an average length of 160 Å (10). Based on x-ray crystallography, the length of FimH is ≈110 Å (22). The length of a tip fibrillum missing FimH (expressed from fimH− mutants) was ≈30 Å (10). The decrease in length of 130 Å in the fimH− mutant is consistent with the loss of no more than one FimH molecule. The 30-Å tip fibrillum in the fimH− mutant is consistent with the presence of one pilin, most likely FimG, because FimG copurifies with FimH as determined by mannose chromatography (10). Thus, if additional FimG and FimH proteins are present in the pilus structure, they presumably would be intercalated into the pilus rod. Thus, the oligomeric complex in the FimDCGH sample may consist primarily of a rod-like structure. It has been suggested previously that occasionally FimH can be intercalated within the type 1 pilus rod (30, 34, 35); the relative quantities of FimG and FimH in the FimDCGH assembly intermediate (Fig. 2A, lane 2) suggest that there may be multiple FimH subunits in the multimer that is formed. If so, it would probably occur by a mechanism other than donor strand exchange.
The coexpression of FimF greatly reduces the multimerization seen in the FimDCGH sample. Presumably, the tail groove of FimF is unable to accommodate the N-terminal extension of FimG but may be specific for the N-terminal extension of FimA. Thus, the N-terminal extension (head) of FimF may drive its assembly to the tail groove of FimG or FimH and preclude further incorporation of FimG molecules. The N-terminal extension of FimA may only be able to bind effectively to the groove of FimF and presumably does not interact efficiently with the tail groove of the FimH adhesin, because strains expressing FimDCAH have very few pili (20, 21) and a very low HA titer (Fig. 1). Thus, although pilins are generally conserved in their head-to-tail contacts (31, 36, 37), there also exists a stereochemical specificity that may help determine the order of pilus assembly. By using such specificity, FimF could function to adapt the FimG–FimH complex to the subsequently added FimA subunits as translocation and assembly proceed.
Over 7 million cases of urinary tract infections are reported annually in the United States, suggesting that some uropathogenic E. coli are able to persist in the bladder despite innate host defense mechanisms. It has been noted that type 1 pili are significantly shorter than normal when attached to bladder cells (3). One hypothesized explanation for this phenomenon was pilus retraction, and another was physical impedance of outward pilus growth by the host cell (3). It has been suggested previously that FimH intercalation would result in relatively weak points in the type 1 pili, which are thought to be prone to breakage (35). Such breakage could also account, in part, for the shortening of pili that has been seen during the early stages of infection (3). Thus, the type 1 pilus architecture could provide an additional mechanism for preventing attached bacteria from being excreted with the dying bladder umbrella cells. After pilus breakage, newly exposed FimH adhesins (ref. 35 and unpublished data) could promote bacterial attachment and invasion of underlying epithelial cells.
Our data show that the structure of the type 1 pilus is determined by the differences between similar pilins (e.g., FimF vs. FimA), by specialized subunit–subunit interactions, and by chaperone–subunit–usher interactions that occur during assembly and translocation across the outer membrane. The resultant pilus architecture may be well suited to adapt to the cascade of events that occur as a consequence of host–pathogen interactions in the bladder. Understanding the type 1 pilus architecture can lead to an understanding of how uropathogenic E. coli sidestep innate exfoliation responses and move into and invade newly exposed transitional epithelium. Elucidating the structural basis of type 1 pilus formation is a step toward understanding general mechanisms important in the formation of diverse fibrous organelles in pathogenic bacteria.
Acknowledgments
We thank Dr. R. John Collier for allowing much of this work to be undertaken physically in his laboratory and Frederic Sauer, David Thanassi, and Stefan Knight for many helpful discussions. This research was funded in part by National Institutes of Health Grants R01DK51406 (to S.J.H.), R01 R37 AI29549 (to S.J.H.), and R01GM55722 (to E.B.), as well as by a grant from the Medical Foundation (to E.B.).
Abbreviation
- HA
hemagglutination
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160070497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160070497
References
- 1.Connell J, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner J, Burlein J, Barren P, Koenig S, Leath S, Jones C H, Hultgren S J. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 3.Mulvey M A, Lopez-Boado Y S, Wilson C L, Roth R, Parks W C, Heuser J, Hultgren S J. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 4.Lee C. Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 5.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pugsley A. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koomey M. Trends Microbiol. 1995;3:409–411. doi: 10.1016/s0966-842x(00)88989-1. [DOI] [PubMed] [Google Scholar]
- 8.Ginocchio C C, Olmsted S, Wells C, Galan J. Cell. 1994;76:717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- 9.Parsot C, Menard R, Gounon P, Sansonetti P J. Mol Microbiol. 1995;16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 10.Jones C H, Pinkner J S, Roth R, Heuser J, Nicholes A, Abraham S, Hultgren S J. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed K A, Clark M A, Booth T A, Hueck C J, Miller S I, Hirst B H, Jepson M A. Infect Immun. 1998;66:2007–2017. doi: 10.1128/iai.66.5.2007-2017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis G R, Tommassen J. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 14.Linderoth N, Simon M, Russel M. Science. 1997;278:1635–1637. doi: 10.1126/science.278.5343.1635. [DOI] [PubMed] [Google Scholar]
- 15.Shevchik V, Robert-Baudouy J, Condemine G. EMBO J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 17.Saulino E T, Thanassi D G, Pinkner J S, Hultgren S J. EMBO J. 1998;17:2177–2185. doi: 10.1093/emboj/17.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marciano D K, Russel M, Simon S M. Science. 1999;284:1516–1519. doi: 10.1126/science.284.5419.1516. [DOI] [PubMed] [Google Scholar]
- 19.Orndorff P E, Falkow S. J Bacteriol. 1984;159:736–744. doi: 10.1128/jb.159.2.736-744.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell P W, Orndorff P E. J Bacteriol. 1992;174:5923–5935. doi: 10.1128/jb.174.18.5923-5935.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klemm P, Christiansen G. Mol Gen Genet. 1987;208:439–445. doi: 10.1007/BF00328136. [DOI] [PubMed] [Google Scholar]
- 22.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren S J, Knight S. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 23.Sauer F, Fütterer K, Pinkner J S, Dodson K, Hultgren S J, Waksman G. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 24.Barnhart M M, Pinkner J S, Soto G E, Sauer F G, Langermann S, Waksman G, Frieden C, Hultgren S J. Proc Natl Acad Sci USA. 2000;97:7709–7714. doi: 10.1073/pnas.130183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klemm P, Christiansen G. Mol Gen Genet. 1990;220:334–338. doi: 10.1007/BF00260505. [DOI] [PubMed] [Google Scholar]
- 26.Hultgren S J, Schwan W R, Schaeffer A J, Duncan J L. Infect Immun. 1986;54:613–620. doi: 10.1128/iai.54.3.613-620.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzman L, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furste J, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 29.Thanassi D G, Saulino E T, Hultgren S J. Curr Opin Microbiol. 1998;1:223–231. doi: 10.1016/s1369-5274(98)80015-5. [DOI] [PubMed] [Google Scholar]
- 30.Abraham S N, Goguen J D, Sun D, Klemm P, Beachey E H. J Bacteriol. 1987;169:5530–5536. doi: 10.1128/jb.169.12.5530-5536.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soto G E, Dodson K W, Ogg D, Liu C, Heuser J, Knight S, Kihlberg J, Jones C H, Hultgren S J. EMBO J. 1998;17:6155–6167. doi: 10.1093/emboj/17.21.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMicheal J C, Ou J T. J Bacteriol. 1979;138:969–975. doi: 10.1128/jb.138.3.969-975.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thanassi D G, Saulino E T, Lombardo M-J, Roth R, Heuser J, Hultgren S J. Proc Natl Acad Sci USA. 1998;95:3146–3151. doi: 10.1073/pnas.95.6.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krogfelt K A, Klemm P. Microb Pathog. 1988;4:231–238. doi: 10.1016/0882-4010(88)90073-3. [DOI] [PubMed] [Google Scholar]
- 35.Ponniah S, Endres R O, Hasty D L, Abraham S N. J Bacteriol. 1991;173:4195–4202. doi: 10.1128/jb.173.13.4195-4202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girardeau J P, Bertin Y. FEBS Lett. 1995;357:103–108. doi: 10.1016/0014-5793(94)01340-7. [DOI] [PubMed] [Google Scholar]
- 37.Bullitt E, Jones C H, Striker R, Soto G, Jacob-Dubuisson F, Pinkner J, Wick M J, Makowski L, Hultgren S J. Proc Natl Acad Sci USA. 1996;93:12890–12895. doi: 10.1073/pnas.93.23.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hultgren S J, Jones C H, Normark S. In: Escherichia coli and Salmonella. Neidhardt F, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2730–2757. [Google Scholar]
- 39.Hultgren S J, Porter T N, Schaeffer A J, Duncan J L. Infect Immun. 1985;50:370–377. doi: 10.1128/iai.50.2.370-377.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]