Abstract
The response regulator DegU is involved in various late-growth developmental processes in Bacillus subtilis, including the production of degradative enzymes and the development of genetic competence. DegU is essential for the expression of the competence transcription factor, encoded by comK. ComK is required for the transcription of genes encoding the DNA uptake and integration machinery, as well as for the transcription of its own gene. We have purified DegU to study its role in the expression of comK, and we demonstrate here that DegU binds specifically to the comK promoter. The binding of the response regulator DegU to a promoter target had not been reported previously. DNase I protection analyses show that the DegU binding site overlaps with the ComK binding site, and gel retardation experiments indicate that DegU strongly stimulates the binding of ComK to the comK promoter. We propose that DegU functions at the initiation of competence development, when ComK concentrations are insufficient to support comK transcription, by facilitating ComK binding to the comK promoter. DegU therefore acts as a priming protein that primes the autostimulatory transcription of comK. Such priming activity adds a function to the class of response regulator proteins.
To respond to environmental changes, bacteria employ elaborate sensory and regulatory systems. Members of the family of two-component regulatory systems play a major role in these sensory processes. These signal transduction systems consist of an autophosphorylating histidine protein kinase, which responds to a specific signal by transferring a phosphoryl group to its cognate response regulator, generally a transcription factor (1). The soil bacterium Bacillus subtilis responds to environmental signals by differentiating into cells competent for genetic transformation. Genetic research has established that the development of competence requires the activity of several response regulators, including DegU.
In a B. subtilis culture the development of competence is typically initiated toward the end of exponential growth, and is optimal in minimal medium with glucose as the carbon source. A sufficiently high cell density is another prerequisite for optimal competence. Environmental signals are interpreted by a complex signal transduction pathway, which ultimately leads to the synthesis of the competence transcription factor, encoded by comK (2). ComK activates the transcription of the late competence operons (comC, -E, -F, and -G), encoding the DNA binding and uptake machinery, as well as the transcription of genes necessary for general recombination, such as recA and addAB. In addition, ComK is required for the expression of its own gene (3–5).
Full induction of ComK requires the response regulator DegU (6, 7). DegU, and the histidine protein kinase DegS, were originally identified as a two-component system involved in the synthesis of degradative enzymes in B. subtilis (8, 9). Certain mutations in degU or degS result in hyperproduction of degradative enzymes (Hy-phenotype), due to increased phosphorylation of DegU. Such mutations are pleiotropic, and, in addition to giving rise to the Hy-phenotype, prevent competence development. On the basis of the observation that hyperphosphorylation of DegU and inactivation of the degS-degU operon decreased competence, whereas inactivation of degS left competence unaffected, it has been suggested that unphosphorylated DegU is required for competence, whereas DegU-P activates the production of degradative enzymes. This suggestion was supported by the observation that the degU146 mutation, with an impaired phosphorylation site, had no effect on competence, but reduced degradative enzyme production (10, 11). DegU contains a helix–turn–helix DNA binding motif and is therefore assumed to exercise its activity at the level of transcription.
During exponential growth, ComK is inactivated by the formation of a protein complex with MecA and the protease ClpCP, leading to the degradation of ComK. This complex is destabilized by ComS, a protein whose expression is cell density dependent (12–14). The release of ComK from the complex protects ComK from degradation and activates comK transcription, thus initiating a positive autoregulatory loop. Mutations in mecA or clpC prevent the proteolytic degradation of ComK, as a result of which dramatically elevated levels of this protein accumulate even during exponential growth. In such mutants the DegU requirement for competence is bypassed, but ComK is still needed for its own synthesis (6, 15). However, genetic studies have indicated that DegU is not involved in the MecA/ClpCP-dependent regulation of ComK, and it has been suggested that DegU exerts its effect on ComK synthesis directly by means of the comK promoter (7). To test this suggestion, we have purified DegU and performed DNA binding experiments with comK promoter fragments. In this paper we demonstrate that DegU is able to bind specifically to the comK promoter, where it stimulates the binding of ComK. We suggest that DegU is needed to prime comK transcription when the ComK concentration is low. Activation by priming of an autostimulatory response is another way by which response regulators stimulate transcription.
Materials and Methods
General Methods and Materials.
All molecular cloning and PCR procedures were carried out by using standard techniques (16, 17). Restriction endonucleases were obtained from either Boehringer Mannheim Biochemicals or New England Biolabs. Labeled nucleotides were from Amersham. Media for growth of Escherichia coli and B. subtilis have been described by Sambrook et al. (17), and Venema et al. (18). B. subtilis chromosomal DNA was purified as described by Venema et al. (18).
Purification of DegU and ComK.
The C-terminal His6-tag fusion to DegU was constructed by PCR cloning using primers DU1 (5′-CGT GGC CCA TGG CTA AAG TAA ACA TTG-3′) and DU2 (5′-ATA AGA TCT CAT TTC TAC CCA GCC-3′) (9). These primers contain restriction sites for NcoI and BglII, respectively (underlined), which were used to clone DegU as a C-terminal His6-tag fusion, under regulation of an isopropyl β-d-thiogalactoside (IPTG)-inducible promoter, in the expression plasmid pQE60 (Qiagen). The resulting plasmid, pQDU, was used to synthesize DegU-His6 fusion protein in E. coli.
To establish that the His6-tag fusion to DegU did not interfere with comK expression and competence development, the last 525 bp of the degU gene with the His6-tag fusion was cloned into pUC18 carrying a kanamycin-resistance cassette for selection in B. subtilis. This construct, pUDU, was used to transform B. subtilis BD1960 [comG:lacZ(amyE) (CmR)], a derivative of IS75 (his leu met) to achieve a Campbell-type integration at the degU locus, which inactivated the resident degU gene and placed the degU-his6 fusion under control of the degU promoter (13). The dependence of expression of comG-lacZ, one of the late competence genes, was used to test whether the C-terminally His6-tag extension was not deleterious to competence development. Kanamycin-resistant colonies were found to have a DegU+ phenotype on 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) plates, and, when tested for competence development, exhibited no decrease in transformation efficiency.
To isolate DegU-His6, 1 liter LB medium supplemented with 100 μg/ml ampicillin and 0.2% glucose was inoculated with an overnight culture of E. coli strain M15 carrying pQDU and pREP4 (Qiagen). The culture was grown to OD600 of 0.9, induced with 1 mM IPTG, and growth was continued for an additional hour at 37°C. Cells were collected by centrifugation and washed in buffer A (20 mM Tris⋅HCl, pH 8/200 mM NaCl). Pellets were frozen and stored at −70°C. Pellets were resuspended in buffer B (20 mM Tris⋅HCl, pH 8/200 mM NaCl/0.25% Tween-20) supplemented with 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and cells were broken with a French press at 20,000 psi (138 MPa), and centrifuged for 20 min at 20,000 rpm (Beckman SW 41 Ti rotor). The supernatant was mixed with 2 ml of Ni-nitrilotriacetate (NTA) resin (Qiagen), equilibrated with buffer B, and stirred on ice for 1 hr. The mixture was loaded onto a chromatography column and washed with several column volumes of buffer B supplemented with 30 mM imidazole, and subsequently with buffer C (20 mM Tris⋅HCl, pH 8/300 mM NaCl). The fusion protein was eluted with a 30–500 mM imidazole gradient in buffer C. To remove contaminating DNA, elution fractions containing the fusion protein were diluted 10-fold in buffer D [20 mM Tris⋅HCl, pH 8/1 mM EDTA/0.5 mM dithiothreitol (DTT)] and loaded onto a DEAE-Sepharose column (Pharmacia) equilibrated with buffer D, and DegU was eluted with a 0–1 M NaCl gradient in buffer D.
ComK was purified as a maltose binding protein (MBP)-ComK fusion protein on an amylose (New England Biolabs) column and separated from MBP by cleavage with protease factor Xa as previously described (3). After cleavage was complete, Factor Xa was inactivated by the addition of 1 mM PMSF. To separate ComK from MBP and DNA, the protein mixture was loaded onto a DEAE-Sepharose column (Pharmacia) equilibrated with buffer D. MBP was eluted from the column with a 0–50 mM Na2SO4 gradient in buffer D, and ComK was eluted with a 0–1 M KCl gradient in buffer D containing 50 mM Na2SO4, subsequently. Fractions were collected, and the Na2SO4 concentration was increased to 100 mM, to prevent precipitation of ComK. Purifications were monitored by SDS/PAGE, and fractions were checked for the absence of contaminating DNA by agarose-gel electrophoreses and ethidium bromide staining. Proteins were divided into aliquots and stored at −70°C.
Gel Retardation Analyses.
Gel retardation analyses were carried out as described by Hamoen et al. (19). The comK promoter region was isolated by PCR using primers K1 (5′-CCG GAA TTC AGA ATC CCC CCA ATG CC-3′) and K2 (5′-CGG GAT CCC AGT CTG TTT TCT GAC TCA TAT T-3′) and B. subtilis 168 chromosomal DNA as template. The resulting 325-bp fragment extends from −255 to +54 relative to the transcriptional start of comK (20), and was end-labeled with T4 polynucleotide kinase and [γ-32P]ATP. Purified protein and approximately 0.05 ng/μl probe were premixed on ice in binding buffer [20 mM Tris⋅HCl pH 8/100 mM KCl/5 mM MgCl2/0.5 mM DTT/10% (vol/vol) glycerol with 0.05% Nonidet P-40 and 0.05 mg/ml BSA], containing 0.05 μg/μl poly(dI-dC) as competitive nonspecific DNA. After 20-min incubation at 37°C, samples were loaded on a nondenaturing 6% polyacrylamide gel. Gels were run in TAE buffer (40 mM Tris acetate, pH 8/2 mM EDTA) at 7 V/cm, using a Bio-Rad-minigel system, dried, and autoradiographed. The fraction of comK promoter fragments, retarded as a consequence of ComK binding, was estimated by film scanning with an LKB ultrascanXL laser densitometer.
Footprinting Analyses.
DNase I footprints were obtained as described by Hamoen et al. (19). The DNA probes were obtained by PCR amplification using primers K1 and K2 which were end-labeled by using [γ-32P]ATP. Binding reactions were performed as for the gel retardation experiments, in a total volume of 40 μl. DNA fragments containing approximately 100,000 cpm were added to each reaction mixture. After 20-min incubation at room temperature, 10 μl of 10 mM CaCl2 and 0.75 unit of DNase I were added, and after 1 min the reactions were terminated with 140 μl of stop solution (192 mM sodium acetate/32 mM EDTA/0.14% SDS/64 μg/ml yeast tRNA). The samples were extracted with phenol/chloroform and precipitated with ethanol. The precipitates were resuspended in 3 μl of loading buffer. The DNase I products were analyzed by electrophoresis on a 6% polyacrylamide/urea gel. The resolution of the DNA sequence was enhanced by applying an electrolyte gradient during electrophoresis (16). Maxam–Gilbert G+A reactions were run with each experiment to locate sequence positions and protected regions (17).
Results
DegU Binds Specifically to the comK Promoter.
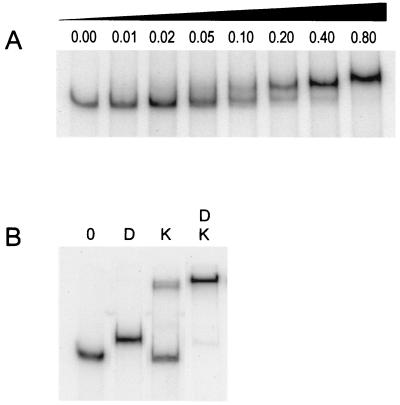
On the basis of genetic analyses, Hahn et al. (7) postulated that the response regulator DegU stimulates comK expression directly at the transcription level. To examine whether the comK promoter is a target for DegU, we purified the response regulator as a C-terminal His6-tag fusion protein. The addition of a C-terminal His6-tag did not interfere with competence development in B. subtilis, indicating that the in vivo activity of DegU is not impaired by the tag. A DNA fragment containing the comK promoter region was incubated with increasing concentrations of purified DegU. As documented in Fig. 1A, the comK promoter contains a specific binding site for DegU, which is supported by the fact that retardation occurs despite the presence of a large excess of nonspecific competing DNA [poly(dI-dC)]. Under the same conditions, DNA fragments containing the recA promoter displayed no DegU binding, although these fragments do bind ComK (4).
Figure 1.
Gel mobility shifts of a 32P-labeled comK promoter fragment. (A) Incubation with increasing concentrations of DegU. Concentrations are given in μM. (B) Effect of DegU (D) on ComK (K)-induced retardation. Protein concentrations used were 0.15 μM and 0.22 μM for DegU and ComK, respectively. The left lane (0) contained no protein.
DegU Enhances ComK Binding.
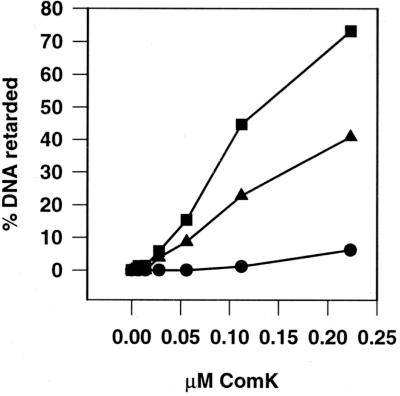
ComK itself activates the transcription of comK. It has previously been shown that ComK binds upstream of the comK transcription initiation site, where it presumably recruits the RNA polymerase (19). Since DegU also binds to the comK promoter, we tested whether DegU might enhance the binding of ComK. As shown in Fig. 1B, when the comK promoter-containing DNA fragment was incubated with both ComK and DegU, a supershift was observed, compared with that caused by incubation with DegU or ComK alone. Apparently, the two proteins are able to bind to this promoter simultaneously. Moreover, the absence of an intermediately migrating band, at the position of the DegU-bound DNA fragment, suggests that DegU stimulates binding of ComK. This idea was substantiated by a gel retardation experiment in which the ComK concentration was varied in the presence or absence of a fixed amount of DegU (Fig. 2). The resulting curves clearly indicate that DegU strongly stimulates ComK-dependent retardation.
Figure 2.
Effect of DegU on the binding of ComK to the comK promoter. The percentage of ComK-retarded DNA fragments is plotted against the ComK concentrations used. The DegU concentrations used were 0 μM (●), 0.08 μM (▴), and 0.15 μM (■).
DegU Binds Between the Two ComK Dimer Binding Sites.
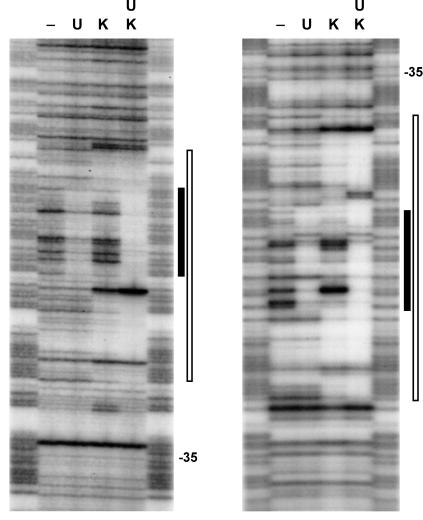
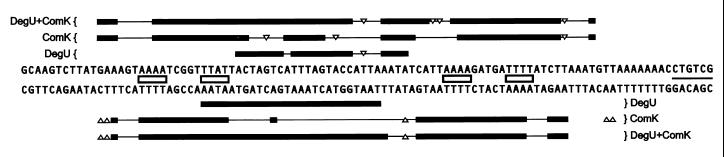
Recently, it has been shown that ComK operates as a double dimer, in which each dimer recognizes a dyad symmetrical consensus sequence (AAAAN5TTTT), the AT-box (19). In the comK promoter, the distance between the two AT-boxes is 44 bp. Because DegU stimulates binding of ComK, it is likely that DegU binds in close proximity to at least one AT-box. To determine the location of DegU binding to the promoter fragment, we performed DNase I protection analyses with 32P-labeled comK promoter fragments in the presence of DegU (Fig. 3). For comparison, DNase I protection analyses were also performed in the joint presence of DegU and ComK, and in the presence of ComK alone. The resulting footprints are summarized in Fig. 4. Figs. 3 and 4 show that DegU protects the DNA region between the two AT-boxes. As the bases protected by DegU and ComK individually are almost the same as those jointly protected by the two proteins, neither of the proteins seems to displace parts of the other when both have bound to the DNA helix.
Figure 3.
DNase I footprinting analysis of the comK promoter region in the presence of DegU, ComK, or both proteins. Left and Right represent the footprints of the upper and lower strands, respectively. DegU (U) and ComK (K) concentrations used were 0.6 μM and 0.9 μM, respectively, and 0.3 μM for both DegU and ComK when jointly present. Footprints are flanked by G+A sequence ladders. The region protected by DegU and ComK are marked with closed and open bars, respectively, and the positions of the −35 promoter sequences are indicated.
Figure 4.
Summary of the DegU and ComK DNase I footprints of the comK promoter. Protected bases are marked by bars and hypersensitive sites by triangles. The ComK-dimer recognition sequences (AT-boxes) are marked with boxes, and the −35 promoter sequence is underlined.
Cooperative Binding Is Not Mediated by Protein–Protein Interactions.
As DegU binds between the two ComK dimers, the facilitation of binding of ComK by DegU may be based on protein–protein interactions. Sogaard-Andersen and Valentin-Hansen (21) have shown that cooperative binding of the E. coli transcription factors CytR and cAMP receptor protein (CRP) to the deoP2 promoter is mediated by protein–protein interactions. When they abolished the CytR binding sequence, located between the two CRP binding sites of the deoP2 promoter, cooperative binding of both proteins was still observed. In contrast, when the DegU binding region between the AT-boxes of the comK promoter was replaced, we observed no stimulation of ComK binding in the presence of DegU. In addition, we performed affinity chromatography experiments with purified MBP-ComK and DegU-His6-tagged fusion proteins, and found that an affinity column loaded with MBP-ComK did not retard the elution of DegU-His6, nor did an affinity column loaded with DegU-His6 retard the elution of ComK (data not shown). These results suggest that the DegU-stimulated binding of ComK is not based solely on protein–protein interactions.
DNA Binding Drugs Stimulate Binding of ComK.
On the basis of the distance between the ComK dimer binding sites, ComK-regulated promoters can be classified into three groups. In the recA and addAB promoters the two AT-boxes are separated by an interval of 21 nucleotides. Assuming 10.5 nucleotides per helical turn, this corresponds to two DNA-helix windings, as a result of which both ComK dimers are located at the same face of the DNA helix. This configuration has been shown to be a requisite for ComK tetramerization (19). The second class of ComK-dependent promoters are those of the late competence genes, comC, -E, -F, and G, in which the AT-boxes are separated by a 31-nucleotide interval, corresponding to three helical turns. The comK promoter is the only known ComK-regulated promoter in which the separation of AT-boxes amounts to four helical turns (44-nucleotide interval). To form a tetramer, the two ComK dimers must span 44 bp. To overcome this distance, it is likely that the DNA helix must bend, and indeed it has been shown that ComK binding is accompanied by substantial bending of the DNA (19). Possibly, DegU stimulates the interaction between the ComK dimers by inducing DNA bending. To determine whether DegU bends the comK promoter, we performed circular permutation analyses (22). However, no substantial bending of the comK promoter fragment was observed (data not shown).
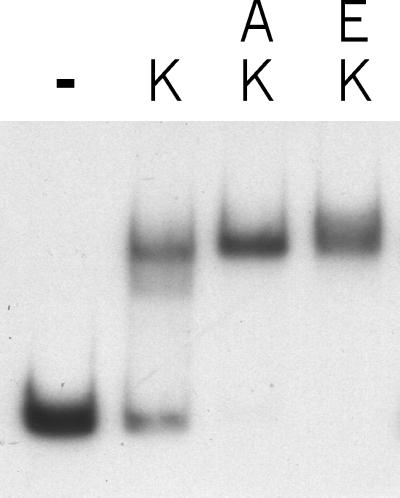
DNA binding experiments in the presence of DNA binding drugs indicated that some of these enhanced binding of ComK. As shown in Fig. 5, a micromolar concentration of actinomycin D stimulated the binding of ComK to the comK promoter. Of the different DNA binding agents tested, ethidium bromide showed a comparable effect (Fig. 5). Actinomycin D as well as ethidium bromide is known to unwind the DNA helix by −26° (23, 24). On the basis of the finding that these drugs simulate the increased binding of ComK by DegU, and since we found no evidence for protein–protein interactions between ComK and DegU, we hypothesize that DegU stimulates ComK binding by altering the shape of the DNA helix between the two AT-boxes, such that contact between the two ComK dimers is facilitated.
Figure 5.
Effect of DNA binding agents on the binding of ComK to the comK promoter. A 32P-labeled comK promoter fragment was incubated with 0.4 μM ComK (K) in the absence (−) or presence of 6 μM actinomycin D (A) or 6 μM ethidium bromide (E). No substantial retardation of the comK promoter fragment was observed when only actinomycin D or ethidium bromide was present (not shown).
Discussion
The response regulator DegU is required for the synthesis of the competence transcription factor encoded by comK. In an attempt to understand the function of DegU in the regulation of competence, we purified the protein and examined whether DegU might be directly involved in the transcriptional control of comK. In this study we show that DegU binds specifically to the comK promoter, which is, to our knowledge, the first reported DNA target for DegU. On the basis of this result we conclude that DegU is directly involved in the transcription of comK. In support of this conclusion we present compelling evidence that DegU stimulates ComK binding to its own promoter.
Synergistic binding of transcription-regulating proteins has been described for other prokaryotic systems, such as deoP2 repression by the E. coli CytR and CRP proteins (21), and repression of the E. coli gal promoter by the concomitant binding of the GalR repressors and the bacterial histone-like protein HU (25). To our knowledge, this is the first report in which a response regulator functions as an accessory protein to stimulate DNA binding of an essential transcription factor.
DNase I protection analyses revealed a 30-bp sequence as the DegU recognition site, located near position −80 relative to the transcription start site. Several DegU mutations result in increased transcription of aprE and sacB and hyperproduction of the enzymes encoded by these genes, subtilisin and levansucrase, respectively (11). Upstream of aprE and sacB, Henner et al. (26) identified similar regions, possibly defining the binding site for DegU∼P. However this inference has never been supported by protein binding studies. Comparison between these regions and the DegU binding site of comK revealed no sequence similarity. Dartois et al. (27) aligned several more promoters whose activity is influenced by degU mutations, and they postulated a putative DegU-binding site with the nucleotide sequence AGAAN11TTCAG. Yet, also in this case, the DegU-binding site of the comK promoter showed no similarity. Centered within the DegU-protected region is a dyad with its arms positioned one turn of the helix apart (TACTAN6TAGTA). As no other DegU-binding DNA sequences have been confirmed by DNA–protein interaction studies, and no mutational analysis has been carried out, the significance of this dyad symmetrical sequence for DegU binding remains to be established.
ComK functions as a tetramer composed of two dimers, each recognizing the motif AAAAN5TTTT, defined as the AT-box. In the comK promoter the two AT-boxes are located 44 bp apart, and footprinting experiments showed that DegU binds between these two ComK dimer binding sites. We found no evidence that cooperative binding of ComK and DegU is based solely on protein–protein interactions, and circular permutation analyses gave no support for a clear DegU-induced bending of the comK promoter, although, because of a relatively poor resolution of the circular permutation analyses, a slight degree of bending might have escaped our attention. However, the stimulating effect of micromolar concentrations of actinomycin D and ethidium bromide on ComK binding indicated that changing the winding angle of the comK promoter does stimulate the formation of ComK tetramers. These intercalating agents unwind the DNA helix by −26°, and unwinding of the DNA helix between the AT-boxes could optimize the phasing of AT-boxes (23, 24). The separation of AT-boxes in other ComK-activated promoters amounts to 21 or 31 nucleotides, almost exactly 2 and 3 helical turns. As a result, both AT-boxes are positioned exactly at the same face of the DNA helix, which is essential for optimal dimer interaction. In the comK promoter the AT-boxes are separated by a 44-nucleotide interval, 2 nucleotides more than in the ideal case of 4 helical turns. As a consequence the AT-boxes are dislocated by about +70° relative to the DNA helix axis. Partial unwinding the DNA could therefore restore optimal phasing of AT-boxes. In conclusion, we propose that DegU does not stimulate ComK binding by means of protein–protein contacts, but by diminishing the physical distance between the ComK dimers, and as such stimulating tetramerization of ComK on the comK promoter. DegU could achieve this by partial unwinding and possibly by slightly bending the DNA helix.
The comK promoter accommodates two additional transcription regulators: AbrB and CodY (L.W.H., M. Marahiel, and P. Serror, unpublished results; ref. 28) (Fig. 6). AbrB represses various starvation-induced differentiation processes in B. subtilis, among which is competence development (29). During the transition to the stationary growth phase levels of AbrB decline. CodY is involved in the nutritional repression of competence development (20). With the conclusion that DegU regulation occurs at the level of comK transcription, the number of proteins acting at the comK promoter totals four. To our knowledge, such complexity has so far not been documented for prokaryotes. The possibility that DegU may act by antagonizing the repression by AbrB and CodY can be excluded on the basis of genetic data (7, 30).
Figure 6.
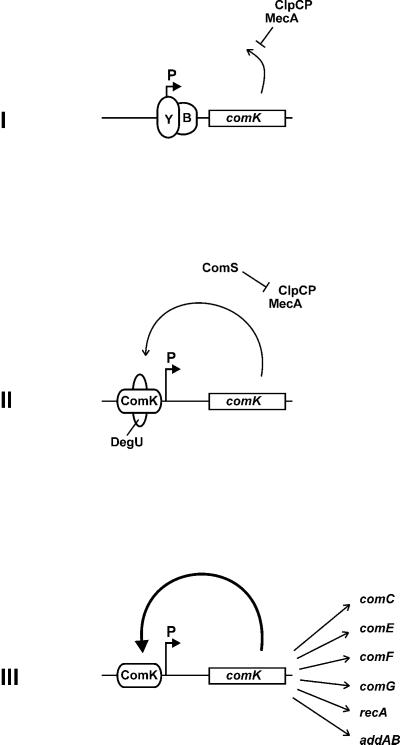
Schematic representation of the transcriptional and posttranscriptional control of comK in competence development. (I) During logarithmic growth, transcription of comK (open box) is repressed by the transcriptional repressors AbrB (B) and CodY (Y). Residual ComK is degraded by the action of MecA and ClpCP. (II) At the beginning of the stationary growth phase, AbrB and CodY repression is raised, and the MecA/ClpCP complex is destabilized by the cell density-induced synthesis of ComS. DegU stimulates binding of ComK, initiating the autostimulatory expression of comK. (III) When ComK concentrations are sufficiently high (bold arrow), ComK activates the expression of genes constituting the DNA-uptake system (comC, -E, -G, -F), of the DNA-integration system (recA, addAB), and of comK itself, independent of DegU. Binding of AbrB, CodY, DegU, and ComK to the comK promoter (P) is indicated with ellipses.
Inactivation of clpC or mecA by mutation strongly induces comK expression. In these mutants, DegU is no longer required for comK expression, yet ComK itself is still indispensable. Apparently, ComK is the main transcriptional activator of comK and, at sufficiently high concentrations, is able to function without DegU. The possibility that DegU functions independently as a transcriptional activator and stimulates the synthesis of low amounts of ComK, sufficient to start the autostimulatory ComK production, can be excluded, because no residual transcription activity from the comK promoter is observed when ComK is inactivated. We therefore propose that the main function of DegU in the development of competence is to stimulate binding of ComK at the onset of competence development, when the small amounts of ComK, released from the ComK/MecA/ClpCP complex by ComS, are insufficient for binding. As such, the response regulator DegU primes the autostimulatory comK transcription loop (Fig. 6) and plays a distinctive role as a regulatory priming protein, potentiating the binding of a transcription activator to its own promoter.
Acknowledgments
We thank Jeanette Hahn and Kursad Turgay for helpful discussions. This work was supported by the Netherlands Organization of Scientific Research (NWO) under the auspices of the Netherlands Foundation for Chemical Research (SON), and by National Institutes of Health Grant GM57720.
Abbreviation
- MBP
maltose binding protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160010597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160010597
References
- 1.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Dubnau D. In: Bacillus subtilis and Other Gram-Positive Bacteria. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 555–584. [Google Scholar]
- 3.van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. Mol Microbiol. 1995;15:455–462. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- 4.Haijema B J, van Sinderen D, Winterling K, Kooistra J, Venema G, Hamoen L W. Mol Microbiol. 1996;22:75–85. doi: 10.1111/j.1365-2958.1996.tb02657.x. [DOI] [PubMed] [Google Scholar]
- 5.Haijema B J, Hamoen L W, Kooistra J, Venema G, van Sinderen D. Mol Microbiol. 1995;15:203–211. doi: 10.1111/j.1365-2958.1995.tb02235.x. [DOI] [PubMed] [Google Scholar]
- 6.van Sinderen D, Venema G. J Bacteriol. 1994;176:5762–5770. doi: 10.1128/jb.176.18.5762-5770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn J, Luttinger A, Dubnau D. Mol Microbiol. 1996;21:763–775. doi: 10.1046/j.1365-2958.1996.371407.x. [DOI] [PubMed] [Google Scholar]
- 8.Henner D J, Yang M, Ferrari E. J Bacteriol. 1988;170:5102–5109. doi: 10.1128/jb.170.11.5102-5109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunst F, Debarbouille M, Msadek T, Young M, Mauel C, Karamata D, Klier A, Rapoport G, Dedonder R. J Bacteriol. 1988;170:5093–5101. doi: 10.1128/jb.170.11.5093-5101.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahl M K, Msadek T, Kunst F, Rapoport G. J Biol Chem. 1992;267:14509–14514. [PubMed] [Google Scholar]
- 11.Msadek T, Kunst F, Rapoport G. In: Bacillus subtilis and Other Gram-Positive Bacteria. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 1993. pp. 555–584. [Google Scholar]
- 12.Solomon J M, Lazazzera B A, Grossman A D. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 13.Turgay K, Hamoen L W, Venema G, Dubnau D. Genes Dev. 1997;11:119–128. doi: 10.1101/gad.11.1.119. [DOI] [PubMed] [Google Scholar]
- 14.Turgay K, Hahn J, Burghoorn J, Dubnau D. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roggiani M, Hahn J, Dubnau D. J Bacteriol. 1990;172:4056–4063. doi: 10.1128/jb.172.7.4056-4063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidham J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1998. [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Venema G, Pritchard R H, Venema-Schroder T. J Bacteriol. 1965;89:1250–1255. doi: 10.1128/jb.89.5.1250-1255.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamoen L W, Van Werkhoven A F, Bijlsma J J, Dubnau D, Venema G. Genes Dev. 1998;12:1539–1550. doi: 10.1101/gad.12.10.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Sinderen D, ten Berge A, Hayema B J, Hamoen L, Venema G. Mol Microbiol. 1994;11:695–703. doi: 10.1111/j.1365-2958.1994.tb00347.x. [DOI] [PubMed] [Google Scholar]
- 21.Søgaard-Anderson L, Valentin-Hansen P. Cell. 1993;75:557–566. doi: 10.1016/0092-8674(93)90389-8. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Zwieb C, Wu C, Adhya S. Gene. 1989;85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- 23.Cons B M, Fox K R. Biochem Biophys Res Commun. 1990;171:1064–1070. doi: 10.1016/0006-291x(90)90792-l. [DOI] [PubMed] [Google Scholar]
- 24.Barcelo F, Muzard G, Mendoza R, Revet B, Roques B P, Le Pecq J B. Biochemistry. 1991;30:4863–4873. doi: 10.1021/bi00234a005. [DOI] [PubMed] [Google Scholar]
- 25.Aki T, Adhya S. EMBO J. 1997;16:3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henner D J, Ferrari E, Perego M, Hoch J A. J Bacteriol. 1988;170:296–300. doi: 10.1128/jb.170.1.296-300.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dartois V, Debarbouille M, Kunst F, Rapoport G. J Bacteriol. 1998;180:1855–1861. doi: 10.1128/jb.180.7.1855-1861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serror P, Sonenshein A L. J Bacteriol. 1996;178:5910–5915. doi: 10.1128/jb.178.20.5910-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strauch M A. In: Bacillus subtilis and Other Gram-Positive Bacteria. Sonenshein A L, Hoch J A, Losick R, editors. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 757–764. [Google Scholar]
- 30.Hahn J, Roggiani M, Dubnau D. J Bacteriol. 1995;177:3601–3605. doi: 10.1128/jb.177.12.3601-3605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]