Abstract
Epstein–Barr virus (EBV) is a ubiquitous human herpesvirus that causes infectious mononucleosis and is etiologically associated with malignancies of multiple origins. EBV enters cells through a cascade of interactions between its envelope glycoprotein gp350 and the gp42-gH-gL complex with cellular receptors. Membrane fusion is catalyzed by the binding of gp42, a member of the C type lectin family, to HLA class II molecule HLA-DR, -DP, or -DQ. Here we demonstrate that only a subset of HLA-DQ alleles mediates EBV entry, indicating that individuals expressing these alleles may offer unique sites for EBV infection and subsequent sequelae. Additionally, the specific site within HLA-DQ determined to be essential for EBV entry is homologous to a site within MHC class I shown by structural studies to bind to the C type-lectin-like natural killer receptor, providing insight into the biochemical nature of the gp42–HLA class II interaction.
Epstein–Barr virus (EBV), also designated human herpesvirus 4 (HHV-4), is one of eight human herpesviruses that establish latent infection in human hosts (1, 2). Infection is prevalent in all human populations, and, along with HHV-8, EBV is the only herpesvirus that plays an etiological role in human malignancies (1, 3). EBV is the causative agent in endemic Burkitt's lymphoma and undifferentiated nasopharyngeal carcinoma (1, 3). In immune-suppressed individuals, the viral infection correlates with a variety of proliferative disorders including oral hairy leukoplakia, immunoblastic lymphomas, and an unusual tumor of muscle origin (1, 3). In addition, EBV may be a factor in some forms of Hodgkin's disease and T cell lymphomas (1, 3).
EBV association with diseases of multiple tissue origins indicates its access into a wide variety of host cells in vivo. However, EBV entry in vitro is restricted largely to B cells. The initial event required for entry into B cells is the interaction of the major viral envelope glycoprotein, gp350, to its receptor CD21 through a sequence similar to that of the C3d component of complement (4, 5). Viral envelope fusion with the host cell membrane requires the additional interaction of the ternary EBV glycoprotein gp85-gp25-gp42 complex with its cellular ligand (6, 7). The HLA class II protein HLA-DR binds to gp42 and can serve as a coreceptor for EBV entry (8, 9). More recent results also illustrate that EBV also can use HLA-DP or HLA-DQ as a coreceptor to mediate entry (10).
HLA class II antigens are α/β heterodimeric cell surface glycoproteins that function to present processed antigens to CD4+ T lymphocytes. The HLA class II region encodes three loci encoding functional HLA class II antigens, HLA-DR, -DP, and -DQ (11). Each isotype is highly polymorphic and encodes many alleles, which creates vast diversity among HLA class II antigens (11). However, individuals express only a small subset of the possible HLA class II alleles. All three isotypes are capable of functioning as a coreceptor for EBV entry; yet, the ability of specific alleles to mediate EBV is not well characterized. If HLA class II alleles demonstrate a differential capability in conferring host cells susceptible to EBV, it may predispose individuals with certain haplotypes to specific sequelae of EBV infection.
Through mutational analysis of different HLA-DQ alleles this report establishes that a glutamic acid at residue 46 of the HLA class II β-chain is necessary for EBV entry, demonstrating that individual HLA class II haplotypes indeed may be important in EBV infection. Although all known HLA-DR and -DP alleles encode a glutamic acid residue at this position, it is unique to only a small subset of HLA-DQ alleles, suggesting a link between individuals expressing these alleles and distinctive pathogeneses upon EBV infection. The domain surrounding residue 46 is homologous to a site on MHC class I that interacts with the murine natural killer (NK) receptor Ly49A (12). Despite amino acid sequence differences, MHC class I and HLA class II molecules structurally are very similar (13, 14). This observation in addition to the fact that Ly49A and gp42 are both members of the C type-lectin-like superfamily indicate these interactions may be structurally similar (8, 15). Therefore, these results provide insight into the biochemical interaction between gp42 and HLA class II molecules.
Materials and Methods
Cell Culture, Transfection, and Infection.
721.174 cells (16) were cultured in RPMI 1640 medium supplemented with 15% newborn calf serum (Sigma) and antibiotics. Transfections were carried out by electroporation using a Gene Pulser (Bio-Rad). Cells (2 × 107) were electroporated in 0.4-cm gap cuvettes (Bio-Rad) at 0.280-kV and 960-μF capacitance. Plasmids containing the specified cDNAs were electroporated at a 1:1(:1) ratio. A total of 50 μg of DNA was used for all electroporations. Twenty-four hours after transfection, 721.174 cells were exposed to 3 × 105 “green” units under constant agitation at 37°C. After exposure to the virus, cells were pelleted and resuspended in fresh medium.
Plasmids and Strains.
The cDNAs encoding HLA-DQ α*0501, α*0301, β*0201, and β*03032 were inserted into pSG5 (Stratagene) as described previously (10). Invariant chain (Ii) p33–143 (17) and Ii p41/43 (18) were excised from pSP46 by BamHI endonuclease, inserted into pSG5, and screened for proper orientation. HLA-DQ β*02–03032 was created by excising the EcoRI–SacI fragment of HLA-DQ β*03032 and replacing it with the same fragment of HLA-DQ β*0201. The reverse procedure was used to create HLA-DQ β*03–0201. The EBfaV-green fluorescent protein (GFP) recombinant EBV was created by inserting the cDNA encoding enhanced GFP (EGFP) under the control of the cytomegalovirus immediate early (IE) promoter carrying neomycin resistance into the coding sequence for LMP2A as described (19). EBfaV-GFP was harvested from the supernatant of stimulated B95 cells carrying EBV–GFP. B95 cells were stimulated by electroporating 2 × 106 cells with 50 μg of a plasmid containing the EBV Z transactivator in a pSG5 backbone. Stimulated cells were grown in RPMI 1640 medium containing the phorbol ester TPA [12-O-tetradecanoylphorbol 13-acetate, 20 ng/ml (Sigma)] and butyric acid [3 mM (Sigma)]. Five days after stimulation, medium was collected and titered for EBfaV-GFP in terms of “green” units. One green unit is the amount of EBfaV-GFP stock necessary to cause one Daudi cell to fluoresce with EGFP.
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed by using QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer protocols. Point mutations were validated both by restriction digest and by sequencing both strands. The 5′ → 3′ primers for each mutation are listed below. 3′ → 5′ primers for each mutation are the reverse complement of its respective 5′ → 3′ primer. Bold type represents mismatched nucleotides; underlined nucleotides comprise the restriction enzyme recognition sequence. Restriction enzymes are in parentheses.
β*0201 EF→VY, 5′-CGACAGCGACGTGGGGGTATACCGGGCGGTGACG-3′ (AccI); β*0201 LL→PP, 5′-CGGGCGGTGACGCCGCTGGGGCCCCCTGCCGCCGAGTAC -3′ (ApaI); β*0201 DI→EV, 5′-GGAACAGCCAGAAGGAGGTCCTGGAGAGGAAACG-3′ (Eco0109 I); β*0201 KA→TE, 5′-GGACATCCTGGAGAGGACCCGGGCGGAGGTGGACAGGG TGTG-3′ (SmaI); β*0201 E→V, 5′-GCTTCGACAGCGACGTGGGCGTCTTCCGGGCGGTGACG-3′ (BsaHI); β*0201 E→Q, 5′-GCTTCGACAGCGACGTGGGGCAATTCCGGGCGGTGACG-3′ (Tsp509I); β*0201 E→K, 5′-CGACAGCGACGTGGGGAAATTCCGGGCGGTGACG-3′ (ApoI); β*0201 E→D, 5′-GACAGCGACGTGGGGGACTTCCGGGCGGTGAC-3′ (BsmFI); β*03032 V→E, 5′-GCTTCGACAGCGACGTCGGGGAGTATCGGGCGGTGACG-3′ (AatII); and β*03032 G→E, 5′-GCTTCGACAGCGACGTCGAGGTGTATCGGGCGGTGACG-3′ (BsaHI).
Flow Cytometry.
Twenty-four hours after exposure to EBfaV-GFP, cells were stained for 15 min at 4°C by using a primary anti-HLA-DQ antibody Ia3 (ICN). Ia3 binding was detected by a 15-min, 4°C incubation with a secondary goat anti-mouse antibody conjugated to allophycocyanin (APC) (Caltag, South San Francisco, CA). Flow cytometry was carried out by using a FACScalibur (Becton Dickinson) flow cytometer class II-associated invarient chain peptide (CLIP) expression was detected by using the mAb CerCLIP (PharMingen) recognized by a goat anti-mouse antibody conjugated to APC. Flow cytometry data were analyzed by using cellquest software (Becton Dickinson).
Results
Differential Ability of Functional HLA-DQ Alleles to Mediate EBV Infection.
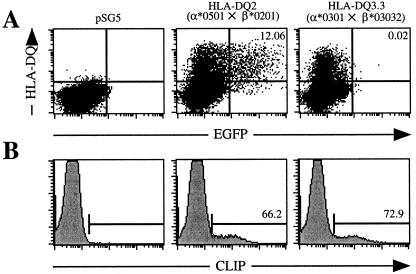
The recent construction of a recombinant EBV reporter virus expressing enhanced GFP, designated EBfaV-GFP (19), allows for the rapid assessment of the cellular receptors required in EBV entry. 721.174 is a CD21+, HLA class II− lymphoblastoid cell line resistant to infection by EBfaV-GFP but is susceptible to EBV infection when transiently transfected with HLA-DR, -DP, or -DQ (10). Although this HLA-DQ molecule is capable of mediating entry, another report has indicated that some HLA-DQ molecules do not bind gp42 and therefore might not be capable of mediating entry (9). Based on amino acid sequence divergence, several different HLA-DQ alleles were screened to determine the extent to which HLA-DQ alleles are capable of mediating EBV entry. Transient expression of different HLA-DQ molecules indicates that HLA-DQ molecules exhibit a contrasting ability to mediate EBV entry (Fig. 1A). Two-color flow cytometry revealed that 721.174 cells transfected with cDNAs encoding HLA-DQ2 (α*0501×β*0201) are susceptible to EBfaV-GFP, as approximately 12% of HLA-DQ2-expressing cells are infected (Fig. 1A). This percentage of infected cells is similar to that of Daudi, a Burkitt's lymphoma cell line (20), which is the cell line most efficiently infected by EBfaV-GFP in culture (10). Alternatively, no 721.174 cells expressing HLA-DQ3.3 molecules (α*0301×β*03032) fluoresce with GFP, demonstrating that HLA-DQ3.3 molecules cannot function as an EBV coreceptor (Fig. 1A). In vector control-transfected cells, no HLA-DQ was expressed and EBfaV-GFP infection was not detectable (Fig. 1A).
Figure 1.
Differential ability of functional HLA-DQ molecules to mediate EBV entry. (A) Susceptibility of 721.174 cells transiently transfected with pSG5, HLA-DQ2, and HLA-DQ3.3 to infection by EBfaV-GFP as analyzed by two-color flow cytometry. Numbers in the upper right-hand quadrants represent the percentage of HLA-DQ-expressing cells infected by EBfaV-GFP. The value for pSG5-transfected cells is >0.00%. Dot plots contain 40,000 events. (B) CLIP expression of pSG5-, HLA-DQ2-, and HLA-DQ3.3-transfected 721.174 cells at the time of infection with EBfaV-GFP. Numbers indicate the percentage of HLA-DQ-expressing cells positive for CLIP expression. Cotransfection of Ii with either HLA-DQ molecule did not enhance CLIP or HLA-DQ expression or alter infection of 721.174 cells with EBfaV-GFP (data not shown). CLIP was not detected on the surface of cells transfected with Ii alone or transfected with the parental plasmid (Fig. 1b and data not shown). Histograms contain 25,000 events.
721.174 cells are deleted for the HLA class II region and lack HLA-DM, which catalyzes peptide loading of HLA class II molecules, but express Ii, which chaperones HLA class II molecules to ensure proper folding and surface expression of the mature HLA class II molecule. Therefore, a majority of mature HLA class II molecules should be associated with CLIP, a peptide cleavage fragment of Ii, bound in the peptide-binding grove. To discount the possibility that DQ3.3 is not functional despite being recognized by an anti-HLA-DQ antibody, CLIP staining on HLA-DQ2- and HLA-DQ3.3-transfected cells was determined (Fig. 1B). On HLA-DQ2-transfected cells, the percentage of cells staining for CLIP was approximately 66.2% of the number of cells that expressed HLA-DQ2 (Fig. 1B and data not shown). CLIP staining on HLA-DQ3.3-transfected cells was measured to be 72.9% of the number of cells that expressed HLA-DQ3.3 (Fig. 1B and data not shown). Detection of similar levels of CLIP on cells transfected with either HLA-DQ molecule indicates that both HLA-DQ molecules were likely to be functional.
N-Terminal 87 aa of HLA Class II β-Chains Contain a Domain Necessary for Entry Activity.
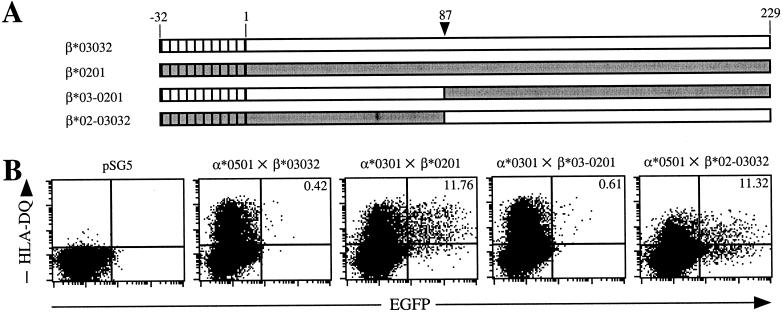
Immunoprecipitation experiments between the HLA-DR and gp42 have indicated that gp42 interacts with the HLA-DR β-chain between amino acids 44 and 108 (8). In addition, the epitopes recognized by HLA-DR antibodies that are capable of blocking EBV entry also map to this region (9). To determine whether the HLA-DQ β-chain was responsible for the entry phenotype, the α-chain of HLA-DQ3.3 (α*0301) was cotransfected with the β-chain of HLA-DQ2 (β*0201). 721.174 cells expressing this hybrid molecule were rendered susceptible to EBfaV-GFP (Fig. 2B, α*0301×β*0201); however, cells expressing the opposite hybrid molecule were resistant to infection (Fig. 2B, α*0501×β*03032), confirming the importance of the β-chain in EBV entry. To define further the portion of the β-chain important for mediating EBV entry, a SacI restriction site common to both HLA-DQ β sequences was used to create two chimeric HLA-DQ β-chains (Fig. 2A). 721.174 cells transfected with HLA-DQ α*0301×β*03–0201 remained resistant to infection (Fig. 2B). In contrast, transfection of HLA-DQ α*0501×β*02–03032 results in efficient entry by EBV. Although a portion of these cells seems susceptible despite an apparent lack of HLA-DQ expression (lower-right quadrant), it is likely that these cells indeed expressed HLA-DQ at the time of infection, because vector control cells are not infectable by EBV. To ensure proper presentation of the hybrid and chimeric molecules, CLIP expression was determined and found to be proportional to HLA-DQ levels at the time of infection (data not shown).
Figure 2.
The N-terminal 87 aa of HLA-DQ β*0201 provide a vital epitope involved in EBV entry. (A) Schematic representation of the HLA-DQ β-chains. Clear regions represent β*03032 amino acid sequence; gray regions represent β*0201 sequence. Hatched portions represent signal peptides. Arrowhead indicates the position of the amino acid encoded at the SacI cleavage site. (B) Susceptibility of 721.174 cells transfected with the indicated HLA-DQ cDNAs to infection by EBfaV-GFP as analyzed by two-color flow cytometry. Numbers in the upper right-hand quadrants represent the percentage of HLA-DQ-expressing cells infected by EBfaV-GFP. The value for pSG5-transfected cells is >0.00%. Dot plots contain 40,000 events.
Glutamic Acid at Residue 46 of HLA-DQ β*0201 Is Necessary for Coreceptor Function.
The ability of the β*02–03032 to mediate infection illustrates that the N-terminal 87 aa of HLA-DQ β*0201 provides a domain that is vital for EBV entry. To gain further insight into the residues responsible for coreceptor activity, the amino acid sequences of HLA-DQ β*0201 and β*03032 as well as the HLA-DR and -DP β-chains previously reported to be capable of mediating EBV entry were analyzed (Fig. 3). Based on the conservation of amino acids among β-chains that mediate EBV entry, we created four double amino acid substitutions in HLA-DQ β*0201 to introduce the corresponding amino acids of HLA-DQ β*03032 (Fig. 3). The EF→VY substitutions at residues 46 and 47 of HLA-DQ β*0201 disrupted coreceptor capability (Fig. 4 and Table 1) whereas the other substitutions did not affect EBV entry (Table 1). The single substitution, E→V at residue 46, produced a phenotype similar to the EF→VY substitution, indicating that a glutamic acid residue at position 46 is crucial for mediating EBV entry (Table 1 and Fig. 4).
Figure 3.
Sequence comparison of the N-terminal 90 (88 HLA-DP β*02012) residues of the mature HLA class II β-chains is shown. Connected arrows indicate the positions of the double amino acid substitutions. Open arrow indicates that a E→V substitution in HLA-DQ β*0201 at position 46 disrupts entry-mediating activity. Solid arrows indicate that substitutions at these positions did not alter entry-mediating activity. Shaded residues, 45 and 46, demonstrate the conservation of glutamic acid among beta alleles that mediate EBV entry. HLA-DR amino acid residues involved in peptide binding are marked with *; barred region indicates the residues involved in the homodimerization of HLA-DR molecules.
Figure 4.
Glutamic acid at residue 46 is necessary for EBV coreceptor activity. Shown is the susceptibility of 721.174 cells transfected with the indicated HLA-DQ β cDNAs to infection by EBfaV-GFP as analyzed by two-color flow cytometry. Mutant HLA-DQ β*0201 alleles were cotransfected with HLA-DQ α*0501; HLA-DQ β*03032 alleles were cotransfected with HLA-DQ α*0301. Numbers in the upper right-hand quadrants represent the percentage of HLA-DQ-expressing cells infected by EBfaV-GFP. The value for pSG5-transfected cells is >0.00%. CLIP expression was determined and found to be proportional to HLA-DQ levels at the time of infection (data not shown). Dot plots contain 40,000 events.
Table 1.
EBV entry-mediating activity of HLA-DQ alleles†
| Allele | Exp. 1 | Exp. 2 | Exp. 3 |
|---|---|---|---|
| α*0301 × β*03032 | 0.00 | 0.02 | 0.09 |
| β*0201 | 8.67 | 11.76 | 11.92 |
| β*03-0201 | 0.22 | 0.61 | 0.48 |
| β*03032 V → E | 13.79 | 13.60 | 9.56 |
| β*03032 G → E | 0.08 | 0.06 | 0.01 |
| α*0501 × β*0201 | 10.19 | 12.06 | 13.16 |
| β*03032 | 0.18 | 0.42 | 0.35 |
| β*02-03032 | 8.44 | 11.32 | 12.20 |
| β*0201 EF → VY | 0.53 | 0.23 | 0.45 |
| β*0201 LL → PP | 14.18 | 13.62 | 13.68 |
| β*0201 DI → EV | 10.82 | 10.56 | 10.33 |
| β*0201 KA → TE | 15.42 | 13.39 | 14.20 |
| β*0201 E → V | 0.61 | 0.58 | 0.39 |
| β*0201 E → Q | 0.98 | 1.17 | 0.44 |
| β*0201 E → K | 0.06 | 0.08 | 0.02 |
| β*0201 E → D | 14.69 | 15.10 | 9.84 |
Numbers represent the percentage of HLA-DQ-expressing cells infected by EBfaV-GFP ad calculated in Fig. 1.
All known HLA-DR and -DP alleles contain glutamic acid at residue 46, providing coreceptor activity for EBV entry in the absence of functional HLA-DQ alleles. This is consistent with the observation that the vast majority of the human population is susceptible to EBV. Based on the crystal structure of HLA-DR and computer models of HLA-DQ, residue 46 is contained within one of the loops connecting two antiparallel β-strands that comprise a portion of the floor of the peptide-binding pocket (14). This site contributes to the top of the cavity between the bottom of the peptide-binding grove and the β2 Ig domain and is homologous to the domain of MHC class I that is important for binding the murine NK cell receptor Ly49A (12). This raises the possibility that the gp42–HLA class II interaction is biochemically similar to the NK receptor–HLA class I interaction.
Only HLA-DQ β*02 Alleles Within the HLA-DQ Locus Can Mediate EBV Entry.
To further characterize the importance of glutamic acid at residue 46, several other substitutions at residue 46 were constructed (Table 1). From these we were able to determine that the negative charge of either glutamic acid or aspartic acid at position 46 was necessary to mediate EBV entry (Table 1 and Fig. 4). A hydrophobic substitution, E→V, the replacement of the carboxylate with an amide group, E→N, or a positively charged substitution, E→K, abolishes the entry-mediating activity of HLA-DQ β*0201 (Table 1 and Fig. 4). To demonstrate the necessity of glutamic acid at amino acid 46, a V→E substitution in HLA-DQ β*03032 was cotransfected with HLA-DQ α*0301 into 721.174 cells. This mutated HLA-DQ3.3 molecule possesses entry-mediating activity equivalent to that of HLA-DQ2 (Figs. 1 and 4 and Table 1). A mAb to gp42, F-2–1 (6), inhibits EBV entry into cells expressing this mutated HLA-DQ3.3 molecule by more than 55%, a result similar to F-2-1 entry inhibition on cells expressing the HLA-DQ2 molecule (data not shown). This indicates that the coreceptor activity exhibited by this mutated HLA-DQ3.3 is dependent on the ability to interact with gp42.
The HLA-DQ β*02 alleles β*0201, β*0202, and β*0203 are the only HLA-DQ β alleles that encode glutamic acid at position 46. Yet, several alleles typified by HLA-DQ β*03011 (Fig. 3) encode a glutamic acid at position 45. To test this phenotype, a G→E substitution at residue 45 in HLA-DQ β*03032 was assayed for the ability to mediate viral entry. β-Chains with this substitution were unable to mediate EBV entry activity (Table 1 and Fig. 4), demonstrating the unique role for HLA-DQ β*02 alleles in HLA-DQ-mediated EBV entry.
Discussion
The pattern of systemic illness during a viral infection depends in large part on the specific host organs or tissues infected and, in many cases, on the capacity of the virus to infect discrete cells within these populations. EBV is associated with diseases of diverse cellular origins, indicating that the virus is able to enter an assortment of tissues. CD21 is expressed on a variety of different cell types in addition to B cells, including certain leukemic T cell lines, immature thymocytes, follicular dendritic cells, and at low levels on normal peripheral blood T cells (21, 22). Studies also have reported expression of CD21 on various epithelial tissues and cell lines (23, 24). Tissue specificity of HLA class II genes typically is restricted to immune cells including B cells, dendritic cells, activated T cells, macrophages, and the thymic epithelium. Transcription of class II genes also may be induced by IFN-γ in many cell types, potentially allowing these cells to present antigens to T cells (25). IFN-γ is produced in response to many viral infections, indicating that the IFN-γ-induced HLA class II expression might allow EBV access into a variety of tissues.
HLA-DQ expression generally is thought to be coordinated with the expression of HLA-DR and HLA-DP (26); however, the discordant expression of HLA-DQ has been reported (27, 28). This observation in conjunction with the known differences in the regulatory regions of HLA-DQ from that of HLA-DR and -DP (29) indicates that HLA-DQ may be differentially expressed in various cell types or tissues. In cell types expressing only HLA-DQ, HLA-DQ β*02+ individuals would be susceptible to EBV infection in sites that are absent in other individuals. Alternatively, HLA-DQ β*02 expression in conjunction with the expression of other HLA-DR or HLA-DP may serve to increase the receptor density at the cell surface so that it is sufficient for efficient EBV entry. In this case, HLA-DQ β*02+ individuals might display unique sites of infection or an increased rate of infection by EBV because of the larger number of functional receptors on cell types expressing multiple HLA class II isotypes.
The prevalence of HLA-DQ β*02 alleles varies by ethnicity, with an average frequency of 20% in Caucasian, 17% in Black, and 9% in Asian populations (11). Further investigation clearly is warranted to determine whether HLA-DQ β*02 gene expression correlates with specific EBV-associated disease progression or other diseases that may have an EBV etiology. Of specific interest is the correlation between insulin-dependent diabetes mellitus (IDDM) and individuals expressing HLA-DQ β*0201. This is the only β allele positively associated with IDDM in Korean populations, and it also is associated with IDDM in Caucasian and Black populations (30, 31). In HLA-DQ β*02-positive individuals, cells that otherwise would not be infected in individuals with other HLA-DQ alleles may become infected with EBV. This could result in the presentation of viral determinants that act as peptide mimics to trigger T cells cross-reactive with diabetes-associated antigens. Alternatively, EBV may infect islet cells, which are capable of expressing HLA class II molecules (32), or cells that may traffic to the pancreas. The ensuing cellular damage and cytokine release resulting from viral infection may lead to an inappropriate immune response, resulting in the destruction of islet cells and subsequent development of IDDM. EBV association with IDDM also would explain the widely divergent age of onset among IDDM patients because onset may correspond temporally with infection.
The mutational analysis described here maps a site within HLA-DQ determined to be essential for EBV entry to a homologous site within MHC class I that is shown by structural studies to bind to the C type-lectin-like NK receptor (12). The high degree of structural homology between MHC class I and HLA class II molecules, in addition to the fact that Ly49A and gp42 are both members of the C type-lectin-like superfamily, indicates that these complexes may be structurally similar. If so, this gives important insights into the nature of gp42-HLA class II-induced membrane fusion. It suggests that like Ly49A, gp42 interacts with HLA class II molecules as a homodimer and also suggests that gp42 interacts with HLA class II molecules in two different ways. First, gp42 may bind HLA class II molecules in trans to a class II molecule on an opposing membrane. Second, gp42 also might interact in cis with a class II molecule in the same membrane.
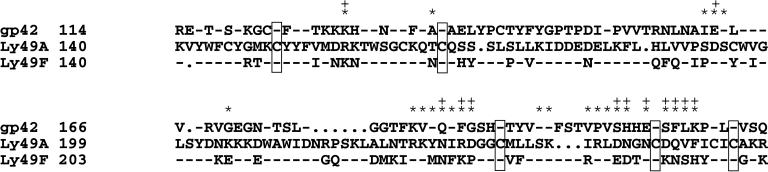
Sequence comparison between the C type-lectin-like domains of gp42 and Ly49A demonstrates only 23% similarity (Fig. 5). However, the residues involved in both cis and trans binding are only weakly conserved. Comparison of these same residues in Ly49F, a close relative of Ly49A, illustrates that they are no more conserved than the same residues in gp42 (Fig. 5). Residue 46 of the HLA-DQ β-chain is a constituent of the site that should provide the cis binding site to gp42, indicating that trans binding is unaffected by residue 46. This raises an interesting possibility that a switch from trans to cis binding triggers fusion between the viral envelope and the host cell membrane.
Figure 5.
Sequence alignment between gp42 and the NK domains of two Ly49 family members. The N-terminal amino acids containing cytoplasmic and transmembrane portions of each protein are omitted. The strictly conserved cysteine residues are denoted by framed boxes. * represent Ly49A amino acids that are involved in cis binding based on the crystal structure of Ly49A complexed with MHC class I. + depicts Ly49A amino acids that comprise the trans-binding domain.
It is striking that EBV may have acquired an NK-like receptor from its host's genome during evolution or, less likely, may have evolved to produce a similar interaction to mediate viral entry. Future structural studies will allow the confirmation of these initial observations and will elucidate the specific mechanisms herpesviruses have acquired to infect the human host. Finally, situations in which individuals demonstrate a decreased or delayed pathogenic outcome in response to microbial or viral agents resulting from the lack of expression of cognate receptors have been reported (33–35). Further investigation will determine whether individuals expressing HLA-DQ β*02 alleles exhibit an increased susceptibility to specific EBV-associated disease progression because of an increase in cellular EBV receptors.
Acknowledgments
We thank the people in the laboratories of R.L. and Dr. P. Spear for providing advice and help. We also thank Drs. E. O. Long, R. M. Hershberg, W. W. Kwok, H. Seifert, P. Spear, S. Miller, and T. S. Jardetzky for the gifts of invaluable reagents and advice. R.L. is a Scholar of the Leukemia Society of America. This work was supported by Public Health Service Grants CA62234 and CA73507 from the National Cancer Institute and Grant DE13127 from the National Institute of Dental and Craniofacial Research. K.M.H. is supported by the training program in the Cellular and Molecular Basis of Disease (T32 GM08061) from the National Institutes of Health.
Abbreviations
- EBV
Epstein–Barr virus
- NK
natural killer
- GFP
green fluorescent protein
- Ii
invariant chain
- CLIP
class II-associated invarient chain peptide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.160171697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.160171697
References
- 1.Rickinson A, Kieff E. In: Fields Virology. Fields B, Knipe D, Howley P, Chanock R, Melnick J, Monath T, Roizman B, Straus S, editors. Vol. 2. Philadelphia: Lippincott; 1996. pp. 2397–2446. [Google Scholar]
- 2.Longnecker R. In: Human Tumor Viruses. McCance D, editor. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 133–172. [Google Scholar]
- 3.Liebowitz D. In: Humor Tumor Viruses. McCance D, editor. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 173–98. [Google Scholar]
- 4.Tanner J, Weis J, Fearon D, Whang Y, Kieff E. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 5.Nemerow G R, Houghten R A, Moore M D, Cooper N R. Cell. 1989;56:369–377. doi: 10.1016/0092-8674(89)90240-7. [DOI] [PubMed] [Google Scholar]
- 6.Li O, Turk S M, Hutt-Fletcher L M. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Hutt-Fletcher L M. J Virol. 1998;72:158–163. doi: 10.1128/jvi.72.1.158-163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spriggs M K, Armitage R J, Comeau M R, Strockbine L, Farrah T, Macduff B, Ulrich D, Alderson M R, Mullberg J, Cohen J I. J Virol. 1996;70:5557–5563. doi: 10.1128/jvi.70.8.5557-5563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Spriggs M K, Kovats S, Turk S M, Comeau M R, Nepom B, Hutt-Fletcher L M. J Virol. 1997;71:4657–4662. doi: 10.1128/jvi.71.6.4657-4662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haan K, Kwok W, Longnecker R, Speck P. J Virol. 2000;74:2451–2454. doi: 10.1128/jvi.74.5.2451-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh S G E, Parham P, Barber L D. The HLA Factsbook. London: Academic; 2000. [Google Scholar]
- 12.Tormo J, Natarajan K, Margulies D H, Mariuzza R A. Nature (London) 1999;402:623–631. doi: 10.1038/45170. [DOI] [PubMed] [Google Scholar]
- 13.Bjorkman P J, Parham P. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 14.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature (London) 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 15.Lanier L L. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 16.DeMars R, Chang C C, Shaw S, Reitnauer P J, Sondel P M. Hum Immunol. 1984;11:77–97. doi: 10.1016/0198-8859(84)90047-8. [DOI] [PubMed] [Google Scholar]
- 17.Strubin M, Long E O, Mach B. Cell. 1986;47:619–625. doi: 10.1016/0092-8674(86)90626-4. [DOI] [PubMed] [Google Scholar]
- 18.Strubin M, Berte C, Mach B. EMBO J. 1986;5:3483–3488. doi: 10.1002/j.1460-2075.1986.tb04673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speck P, Kline K A, Cheresh P, Longnecker R. J Gen Virol. 1999;80:2193–2203. doi: 10.1099/0022-1317-80-8-2193. [DOI] [PubMed] [Google Scholar]
- 20.Klein G, Clements G, Zeuthen J, Westman A. Int J Cancer. 1976;17:715–724. doi: 10.1002/ijc.2910170605. [DOI] [PubMed] [Google Scholar]
- 21.Tsoukas C D, Lambris J D. Immunol Today. 1993;14:56–59. doi: 10.1016/0167-5699(93)90059-T. [DOI] [PubMed] [Google Scholar]
- 22.Reynes M, Aubert J P, Cohen J H, Audouin J, Tricottet V, Diebold J, Kazatchkine M D. J Immunol. 1985;135:2687–2694. [PubMed] [Google Scholar]
- 23.Young L S, Dawson C W, Brown K W, Rickinson A B. Int J Cancer. 1989;43:786–794. doi: 10.1002/ijc.2910430508. [DOI] [PubMed] [Google Scholar]
- 24.Birkenbach M, Tong X, Bradbury L E, Tedder T F, Kieff E. J Exp Med. 1992;176:1405–1414. doi: 10.1084/jem.176.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins T, Korman A J, Wake C T, Boss J M, Kappes D J, Fiers W, Ault K A, Gimbrone M A, Jr, Strominger J L, Pober J S. Proc Natl Acad Sci USA. 1984;81:4917–4921. doi: 10.1073/pnas.81.15.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guardiola J, Maffei A. Crit Rev Immunol. 1993;13:247–268. [PubMed] [Google Scholar]
- 27.Falkenburg J H, Fibbe W E, Goselink H M, Van Rood J J, Jansen J. J Exp Med. 1985;162:1359–1369. doi: 10.1084/jem.162.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ono S J, Bazil V, Sugawara M, Strominger J L. J Exp Med. 1991;173:629–637. doi: 10.1084/jem.173.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersen L C, Beaty J S, Nettles J W, Seyfried C E, Nepom G T, Nepoom B S. J Exp Med. 1991;173:181–192. doi: 10.1084/jem.173.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park Y S, Wang C Y, Ko K W, Yang S W, Park M, Yang M C, She J X. Hum Immunol. 1998;59:794–801. doi: 10.1016/s0198-8859(98)00076-7. [DOI] [PubMed] [Google Scholar]
- 31.She J X. Immunol Today. 1996;17:323–329. doi: 10.1016/0167-5699(96)10014-1. [DOI] [PubMed] [Google Scholar]
- 32.Pujol-Borrell R, Todd I, Doshi M, Bottazzo G F, Sutton R, Gray D, Adolf G R, Feldmann M. Nature (London) 1987;326:304–306. doi: 10.1038/326304a0. [DOI] [PubMed] [Google Scholar]
- 33.Miller L H, Mason S J, Clyde D F, McGinniss M H. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 34.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 35.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]