Abstract
Physiological properties of central nervous system neurons infected with a pseudorabies virus were examined in vitro by using whole-cell patch-clamp techniques. A strain of pseudorabies virus (PRV 152) isogenic with the Bartha strain of PRV was constructed to express an enhanced green fluorescent protein (EGFP) from the human cytomegalovirus immediate early promoter. Unilateral PRV 152 injections into the vitreous body of the hamster eye transsynaptically infected a restricted set of retinorecipient neurons including neurons in the hypothalamic suprachiasmatic nucleus (SCN) and the intergeniculate leaflet (IGL) of the thalamus. Retinorecipient SCN neurons were identified in tissue slices prepared for in vitro electrophysiological analysis by their expression of EGFP. At longer postinjection times, retinal ganglion cells in the contralateral eye also expressed EGFP, becoming infected after transsynaptic uptake and retrograde transport from infected retinorecipient neurons. Retinal ganglion cells that expressed EGFP were easily identified in retinal whole mounts viewed under epifluorescence. Whole-cell patch-clamp recordings revealed that the physiological properties of PRV 152-infected SCN neurons were within the range of properties observed in noninfected SCN neurons. Physiological properties of retinal ganglion cells also appeared normal. The results suggest that PRV 152 is a powerful tool for the transsynaptic labeling of neurons in defined central nervous system circuits that allows neurons to be identified in vitro by their expression of EGFP, analyzed electrophysiologically, and described in morphological detail.
Keywords: PRV-Bartha, PRV 152, suprachiasmatic nucleus, retinal ganglion cell
The use of neurotropic alphaherpesviruses has greatly advanced our ability to visualize ensembles of neurons that contribute to multisynaptic circuits in the central nervous system (CNS) (1). In particular, the attenuated vaccine strain of pseudorabies virus (PRV-Bartha) has been used successfully as a self-amplifying neural tracer after peripheral application or direct injection into brain parenchyma (2–4). The usefulness of PRV as a neural tracer relies on its ability to infect chains of hierarchically connected neurons via specific transsynaptic passage of progeny virus rather than infection by lytic release into the extracellular space (4, 5). Typically, PRV infects the CNS by invading neurons in the periphery and then replicating and spreading to the CNS via synaptically linked neurons. However, PRV can also invade neurons through their somata if the viral concentration is sufficient (6), as evidenced by primary infection of retinal ganglion cells (RGCs) after intravitreal injection of PRV (7–9). Infection of RGCs with PRV-Bartha, followed by viral replication, results in the anterograde transsynaptic infection of a restricted set of retinorecipient neurons [i.e., suprachiasmatic nucleus (SCN), intergeniculate leaflet (IGL), pretectum (PT), and lateral terminal nucleus]. Intravitreal injection of the wild-type virus, PRV-Becker, produces transneuronal infection of neurons in all retinorecipient subcortical regions (7). The factors that determine the specificity of PRV-Bartha infection of selective retinorecipient targets are not completely understood, although deletion of specific genes in PRV-Becker results in a restricted neurotropism identical to that demonstrated with PRV-Bartha (10).
Although viral transsynaptic tracing represents an important methodological advance for the analysis of CNS circuits, functional analysis of virus-infected neurons has been limited to sensory or sympathetic ganglia in culture (11, 12) because of the inability to identify virus-infected neurons in situ. Analyses of electrophysiological properties of neurons, in the context of known functional connections of the recorded neuron, would represent a further important methodological advance for the analysis of CNS circuits.
The development of retrogradely transported fluorescent tracers has allowed investigators to examine the physiology of neurons with known projections (13–15). However, such studies usually require direct and accurate injection of a target region followed by retrograde transport to identify first-order neurons projecting to the target. Other “prelabeling” studies have used constructs of green fluorescent protein to label neurons that possess a particular genetic phenotype, such as expression of gonadotropin-releasing hormone (16). Both of these techniques have allowed examination of the physiological properties of neurons in vitro that possess presumed anatomical or functional correlates in the intact animal.
We now report a neuron-labeling method that combines anatomical prelabeling techniques with transsynaptic labeling methodology to allow physiological analyses of neurons selected for their synaptic connectivity with a target structure, several synapses removed. By using a strain of PRV (PRV 152) isogenic with PRV-Bartha, constructed to express an enhanced green fluorescent protein (EGFP), we tested the hypothesis that transsynaptically labeled neurons infected after intraocular injection of PRV 152 would retain their synaptic connectivity. Our results suggest that second-order infected neurons in the hypothalamic SCN and contralateral RGCs with tertiary labeling from infected retinorecipient neurons receiving bilateral retinal input retain synaptic connectivity while simultaneously robustly expressing EGFP. We conclude that this method will allow physiological examination of a variety of previously intractable functional multisynaptic pathways in the brain.
Methods
Construction of PRV 152.
PRV 152 was constructed by homologous recombination between a plasmid containing an EGFP expression cassette cloned into the middle of the PRV gG gene and the PRV-Bartha genome. Briefly, a 2.6-kbp SalI fragment from PRV-Becker containing the 3′ end of the Us3 gene, the entire gG gene, and the 5′ end of the gD gene, was cloned into the SalI site of pBB3 [a modified pGEM-5zf(+) derivative in which the PstI site and NotI site have been deleted], to generate pBB4. Next, a 2.3-kbp NsiI fragment from pEGFP-N1 (CLONTECH) containing the cytomegalovirus immediate early promoter, EGFP sequences, and a simian virus 40 poly(A) signal, was cloned into a unique PstI site in pBB4 to generate pII1. This leaves about 840 bp of PRV sequence upstream of the EGFP expression cassette and 1,750 bp downstream of the EGFP expression cassette available for homologous recombination with the viral genome. The plasmid pII1 was digested with SalI and cotransfected with purified PRV-Bartha DNA into PK15 cells (a swine kidney cell line). Virus produced after cotransfection was plated on PK15 cells and plaques expressing EGFP were identified with the aid of an inverted epifluorescence microscope. Virus was isolated from EGFP-expressing plaques and subjected to three rounds of purification. Southern blot analysis with the SalI fragment from pBB4 as a probe was performed to verify that the EGFP expression cassette had been properly recombined into the PRV genome (data not shown).
Intraocular Injection of PRV 152.
Syrian hamsters (Mesocricetus auratus; male; >4 months old; LAK:LVG (SYR); Charles River Breeding Laboratories) were maintained under a light/dark cycle of 14 h light/10 h dark with food and water available ad libitum.
Under deep sodium pentobarbital anesthesia (80 mg/kg, i.p.), animals received a unilateral intravitreal injection of 2 μl of 1 × 108 plaque-forming units/ml PRV 152 over a 1-min interval by using a 10-μl Hamilton syringe fitted with a 26-gauge needle; the needle was left in place for an additional 4 min before removing it from the eye. A fresh stock of virus was thawed for each injection. Animals were maintained in a biosafety level 2 laboratory for up to 120 h after injection.
Brain Slice and Retinal Whole-Mount Preparation.
Animals were decapitated under deep sodium pentobarbital anesthesia (100 mg/kg). For acute hypothalamic slices, the brains were rapidly removed from the skull and immersed in ice-cold, oxygenated (95% O2/5% CO2) artificial cerebrospinal fluid (ACSF) containing (in mM): 124 NaCl, 3 KCl, 26 NaHCO3, 1.4 NaH2PO4, 11 glucose, 1.3 CaCl2, 1.3 MgCl2, pH = 7.3–7.4, with an osmolality of 290–305 mosmol/kg. Brains were blocked and mounted on a vibrating microtome, and horizontal slices (400 μm) of the ventral hypothalamus containing the SCN and the proximal optic nerves were prepared, as described (17). Slices were transferred to an interface-style recording chamber, and allowed to equilibrate while continuously perfused with warmed (32–34°C) and oxygenated ACSF for 1–2 h before whole-cell recording. For retinal recordings, the eye contralateral to the PRV 152 injection was removed and cut along the ora serrata, and the intact retina was dissected free from the underlying pigment epithelium in the eyecup while submerged in ice-cold Ames medium (Sigma). The retina was transferred to a submersion-style recording chamber mounted on an upright, fixed-stage microscope equipped for epifluorescence.
Whole-Cell Patch-Clamp Recording.
Recordings of synaptic activity were made by using patch pipettes constructed of borosilicate glass capillaries as described (17). Pipettes were filled with (in mM): 130 potassium gluconate, 1 NaCl, 5 EGTA, 10 Hepes, 1 MgCl2, 1 CaCl2, 3 KOH, 2–4 Mg-ATP and 0.2% biocytin (Sigma), pH 7.2. Noninjected hamsters were used as controls for comparing SCN neuron synaptic activity with PRV 152-injected animals. Whole-cell signals were recorded by using an Axopatch 1D amplifier (Axon Instruments, Foster City, CA). All signals were low-pass filtered at 2–5 kHz, digitized at 44 kHz (Neuro-corder, Neurodata Instruments, New York), and stored on videotape. Electrical stimulation of the optic nerve was performed by using a stimulating electrode made from a twisted pair of Teflon-coated platinum-iridium wires (75-μm diameter) inside a blunted glass micropipette placed over the nerve. Spontaneous inhibitory postsynaptic current (IPSC) and excitatory postsynaptic current (EPSC) frequency were determined over a period of 1–2 min. SCN recordings were done “blind,” and determination of whether the recorded neuron was EGFP expressing was done post hoc by visualizing EGFP fluorescence and biocytin (detected with an avidin-rhodamine conjugate) in the recorded cell. Recordings in retinal ganglion cells were made in a similar manner, except that EGFP-expressing ganglion cells were visualized in living tissue under epifluorescence by using an FITC filter set. Once the cell of interest was targeted, bright-field optics were used to guide the recording pipette onto the cell. Recording from the targeted neuron was verified by briefly observing EGFP-labeled membrane in the tip of the recording pipette and also by post hoc visualization of avidin-rhodamine conjugate.
Hypothalamic Slice and Retinal Histology.
Slices and retinae were fixed in 4% paraformaldehyde and processed to visualize biocytin-filled neurons with an avidin-rhodamine conjugate (Vector Laboratories) as described (17). Tissue was examined on a Leica Aristoplan microscope equipped for epifluorescence, and digital images were captured with a cooled charge-coupled device camera (Spot, Diagnostic Instruments, Sterling Heights, MI).
Results
Transsynaptic EGFP Labeling.
Primary PRV 152 infection of retinal ganglion cells in the injected eye produced transsynaptic labeling in several retinorecipient nuclei, including the SCN and IGL (Figs. 1 and 2). Components of the primary visual system (i.e., dorsal lateral geniculate nucleus and superior colliculus) were not labeled after intraocular PRV 152 injection in the hamster, similar to findings after intraocular PRV-Bartha injection in the rat and mouse (7–9). At longer postinjection intervals (116–120 h), RGCs in the retina contralateral to the injection were also labeled by retrograde transport from infected retinorecipient neurons receiving a bilateral retinal input (Fig. 1). As described for intraocular PRV injections (18), neurons putatively involved in autonomic regulation were also infected (Fig. 1); labeled neurons were observed in select regions of the hypothalamus (e.g., paraventricular nucleus) and the brainstem (e.g., solitary complex). For this study, we concentrated on electrophysiological characteristics of transsynaptically labeled neurons in the SCN and retina, with several recordings also made in the IGL and solitary complex.
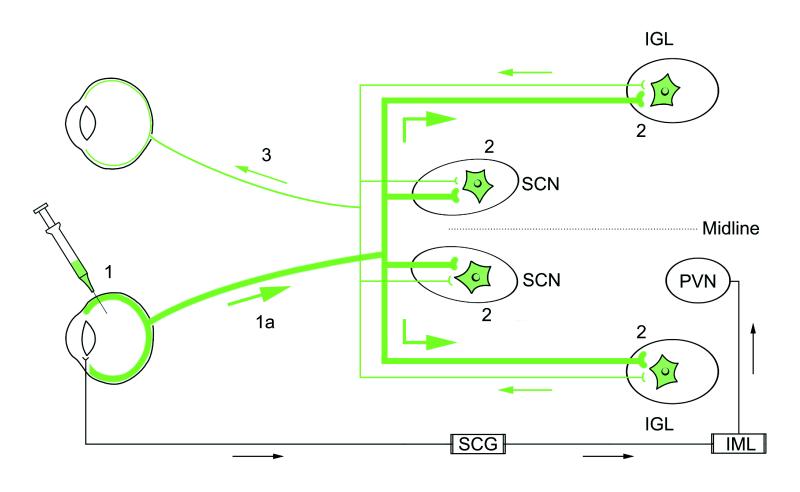
Figure 1.
A schematic diagram illustrating the neuronal circuits labeled after the intraocular injection of PRV 152. Primary infection of retinal ganglion cells occurs after intravitreal injection of PRV 152 (step 1). Viral replication and anterograde transport of virus (step 1a, thick green line) in axons of RGCs follows primary infection. Transsynaptic passage of virus from RGC axons produces secondary infection in a restricted set of retinorecipient central nuclei [i.e., SCN, IGL (shown), PT, and lateral terminal nucleus (not shown) as indicated by green (EGFP-labeled) neurons (2)]. Axon terminals of retinal ganglion cells in the contralateral retina, presynaptic to PRV 152-infected neurons in the SCN, IGL, PT, and lateral terminal nucleus, take up the virus via transsynaptic passage. The virus is retrogradely transported (thin green line) to the contralateral retina, where PRV 152-expressing ganglion cells are easily identified in whole-mount preparations (step 3). Intravitreal PRV 152 injection also infects autonomic afferents to the eye, resulting in retrograde transport of virus to the superior cervical ganglion (SCG), followed by transsynaptic uptake and retrograde transport to preganglionic neurons in the intermediolateral nucleus (IML) of the spinal cord (18). Neurons in the hypothalamic paraventricular nucleus (PVN) are subsequently infected after transsynaptic uptake of the virus from infected IML neurons and retrograde transport.
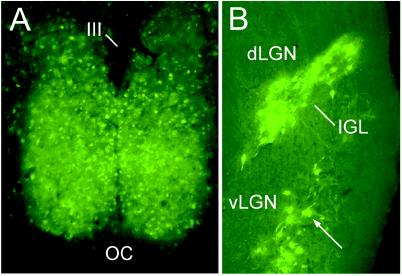
Figure 2.
Micrographs demonstrating transsynaptic EGFP-neuronal labeling after unilateral intravitreal injection of PRV 152 116 h after inoculation. (A) Bilateral transsynaptic infection was evident in the retinorecipient hypothalamic SCN. (B) Bilateral PRV 152 transsynaptic labeling was also evident in the retinorecipient IGL and the ventral lateral geniculate nucleus (vLGN), but not the dorsal LGN. OC, optic chiasm; III, third ventricle.
Synaptic Input to SCN Neurons.
Whole-cell voltage-clamp recordings of synaptic currents were made in 11 SCN neurons from five hamsters injected intraocularly with PRV 152 (Fig. 3). Of these, six neurons were confirmed on histological analysis to have been infected (i.e., they received retinal input from the injected eye), three were uninfected, and two were not recovered (i.e., biocytin was not visualized in the cell). The resting membrane potential (42 ± 6 mV) and input resistance (582 ± 91 MΩ) of infected SCN neurons were similar to those observed in SCN neurons from noninjected hamsters (48 ± 2 mV; 672 ± 61 MΩ; n = 20; P > 0.05). In control animals, electrical stimulation of the optic nerve resulted in an EPSC in ≈55% of neurons. A similar response to optic nerve stimulation was observed in five of six EGFP-labeled neurons (83%); one of three noninfected neurons (33%), and one of two unidentified cells (50%) from PRV 152-injected animals responded with an EPSC. Evoked EPSCs had a poststimulus latency of 7–10 ms, amplitude from a holding potential near −50 mV of −13 to −23 pA, and monoexponential decay time (τ = 3.7–5.0 ms). These results indicated that transsynaptically infected SCN neurons retained their major excitatory input from the retina.
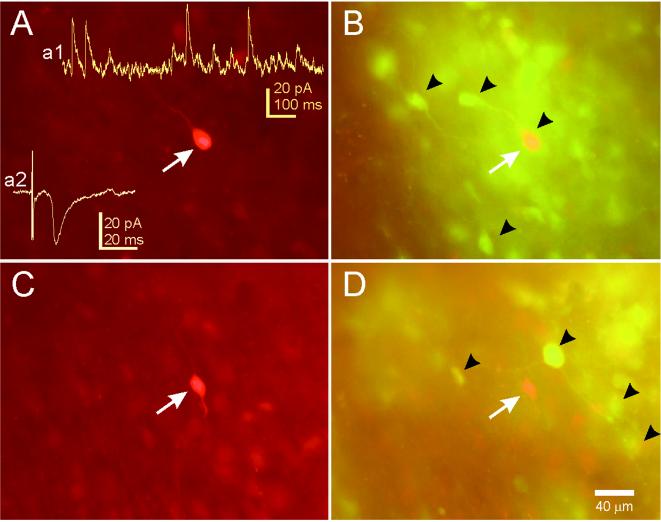
Figure 3.
Examples of neurons recorded in the SCN after PRV 152 intravitreal injection. (A) This SCN neuron was filled with biocytin during a whole-cell patch-clamp recording and visualized with an avidin-rhodamine conjugate. (Inset a1) An example of spontaneous IPSCs in this neuron while voltage clamped at −15 mV. (Inset a2) An example of an EPSC evoked in this neuron by stimulation of the optic nerve. (B) PRV 152 labeling in the same tissue viewed under optics that permits visualization of rhodamine and EGFP concurrently (double-labeled cells appear yellow-orange). The arrow indicates the filled cell in A was also infected with PRV 152. Arrowheads point to examples of EGFP-labeled SCN neurons in the hypothalamic slice. (C) Another biocytin-filled and recorded neuron in the contralateral SCN from the same animal. (D) PRV 152 labeling in the same tissue. The arrow indicates the position of the filled cell in C, which was not infected. Arrowheads point to examples of nearby EGFP-labeled SCN neurons.
Most SCN neurons receive robust local inhibitory input (19). We examined spontaneous IPSCs in PRV-EGFP-labeled SCN neurons to determine if inhibition was also intact in these cells (Fig. 3). Spontaneous IPSCs were observed in all SCN neurons voltage-clamped positive to −50 mV (six infected neurons and six neurons selected at random from noninjected control animals). The spontaneous IPSC frequency was not different between the two groups (19 ± 7 Hz control; 25 ± 5 Hz infected; P > 0.5). Inhibitory input to infected SCN neurons was qualitatively and quantitatively similar to comparable neurons in noninjected animals.
Although less frequent than spontaneous IPSCs, spontaneous EPSCs were also observed in all neurons. The average frequency for spontaneous EPSCs in SCN neurons from control animals was 2.7 ± 1.1 Hz (range 0.3–6 Hz). For infected neurons, the average spontaneous EPSC frequency was 2.4 ± 0.7 Hz (range 0.3–4 Hz). There was no difference in spontaneous EPSC frequency between the two groups (P > 0.5).
Targeted Retinal Ganglion Cells.
Retinal ganglion cells in the contralateral retina, retrogradely labeled from infected retinorecipient neurons, were visually targeted and recorded in a retinal whole-mount configuration (n = 5). Neurons expressing EGFP were easily observed in the RGC layer (Fig. 4). Resting membrane potential was −52 ± 6 mV; input resistance was 297 ± 26 MΩ. In these neurons, spontaneous EPSCs were predominant, with occasional IPSCs (<2 Hz) observed when the neuron was voltage-clamped at a holding potential of −10 to 0 mV in two cells. These results confirmed that retrogradely infected RGCs retained synaptic connectivity and could be targeted in situ for physiological analysis.
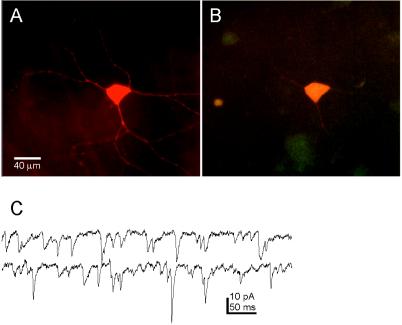
Figure 4.
Biocytin-filled EGFP-expressing retinal ganglion cell viewed in whole mount. (A) Intravitreal PRV 152 injection results in EGFP-expressing ganglion cells in the contralateral retina. This neuron was targeted for recording, filled with biocytin, and reacted with avidin-rhodamine. (B) The same neuron as in A viewed with optics demonstrating double labeling (i.e., EGFP and rhodamine fluorescence). (C) Examples of spontaneous EPSCs recorded from this retinal ganglion cell. Holding potential = −65 mV; traces are continuous.
Discussion
These results describe the synaptic physiology of central neurons that were transsynaptically labeled from a specific target. The EGFP-labeled cells were visually targeted for in vitro electrophysiology, and the infected neurons had intact synaptic inputs within the normal range. Over the past few years, the attenuated PRV-Bartha has emerged as a powerful tool for delineating multisynaptic circuits in the CNS. The wide use of PRV-Bartha is derived from an increased understanding of its life cycle, coupled with compelling evidence that transneuronal passage of PRV is via a synaptic route (1–4, 18, 20). Usually, the virus is detected by using immunocytochemical procedures, providing reliable and reproducible descriptions of multisynaptic CNS circuits. The virus used in this study (PRV 152) was constructed to express EGFP to allow visualization of infected neurons in brain slices acutely prepared for in vitro electrophysiological analyses or in fixed tissue. Similar PRV-Bartha-EGFP constructs have recently been reported and used successfully to trace transneuronal pathways from the periphery to the SCN (21, 22). The EGFP diffuses throughout the cell, which labels neurons sufficiently to be visualized for targeted recordings (16), and also allows identification of dendritic morphology. By combining these two sensitive and powerful techniques, we have demonstrated that functionally related neurons can be analyzed in the context of their local synaptic network.
Synaptic Connectivity of SCN Neurons.
Synaptic input to transsynaptically infected SCN neurons was similar qualitatively and quantitatively to uninfected SCN neurons. PRV 152-infected SCN neurons received robust spontaneous inhibitory input and also excitatory input evoked from optic nerve stimulation. It is likely that the stimulated afferent fibers in the optic nerve were also infected retrogradely from the SCN neuron(s) they contacted, because RGCs in the contralateral eye were infected at the relatively late stage of infection we used. Infection with PRV 152, therefore, does not appear to interfere with normal synaptic function in retinal axons that project to the SCN circadian oscillator.
Quantitatively, at least in terms of frequency, spontaneous EPSCs and IPSCs were indistinguishable between the two groups of infected and uninfected SCN neurons. The range of values for spontaneous postsynaptic current frequency was great for both groups, and only large differences would have been detected. However, the results of these studies indicate that both pre- and postsynaptic elements remain viable in infected animals. Although subtle differences may eventually be found to influence synaptic responses in vivo, the essential connectivity of the system appears to be intact and functional in the infected system. In a few recordings, we also observed spontaneous IPSCs and spontaneous EPSCs in infected neurons from the IGL and nucleus tractus solitarius (data not shown). We used primarily retinorecipient SCN neurons and RGCs to demonstrate the utility of the technique in this study, but analysis of other functional systems should be just as efficient with this method.
Synaptic Connectivity of RGCs.
Consistent with previous reports with PRV-Bartha (7–9), transsynaptically labeled neurons were observed in the SCN and IGL after intravitreal injection of PRV 152. After long postinjection intervals (i.e., ≈120 h), RGCs were retrogradely infected. At least some of these labeled RGCs were retrogradely infected from the SCN and IGL, components of the circadian timing system in the hamster that receive a bilateral retinal input (23).
The EGFP label was sufficiently intense to visualize ganglion cells in retinal whole mounts, allowing identification of these cells for electrophysiological recording. The infected RGCs received robust excitatory synaptic connectivity, similar to a report from whole-cell recordings in mouse ganglion cells in a retinal slice preparation (24). In retinal slice preparations from a variety of species (24–27), strong spontaneous inhibitory synaptic input was also detected in most RGCs. Relative to EPSCs, few IPSCs were observed in PRV 152-infected RGCs. Events were not quantified because of the relatively small number of neurons recorded. The relative paucity of IPSCs, even at relatively depolarized holding potentials (data not shown), suggests that the RGCs that were recorded in this study received little inhibitory input relative to excitatory drive. It is unlikely that this difference resulted from an alteration in synaptic connectivity caused by infection or the EGFP, because spontaneous IPSCs were readily observed in the SCN of similarly treated animals. Perhaps the RGCs retrogradely labeled from the restricted subset of retinorecipient nuclei transsynaptically infected (i.e., SCN, IGL, PT, and lateral terminal nucleus) receive predominately excitatory input.
Current evidence suggests that inhibitory input to ganglion cells arises from GABAergic and glycinergic amacrine cells, whereas excitatory input is from glutamatergic bipolar cells (28). The ratio of EPSCs to IPSCs varies among different ganglion cells (24), and ganglion cells demonstrating sustained responses to light apparently receive a higher ratio of bipolar to amacrine cell input compared with ganglion cells with transient responses (29, 30). Ganglion cells afferent to the SCN demonstrate sustained responses to retinal illumination (31), as do SCN, IGL, and PT neurons (32, 33). Thus, the predominately excitatory input described in the PRV 152-infected RGCs may be characteristic of this subset of cells. However, a greater and more diverse sample will be required to determine if this is the case. The approach described here may allow such an assessment to be made.
The use of EGFP-PRV 152 to examine synaptic connectivity in functionally defined brain regions is a potentially important tool for studying local circuits in the brain. We examined synaptic activity in the SCN and RGCs, but the technique is applicable to a number of systems that are amenable to study with transsynaptic labels. Intraocular PRV injection has been used to define central presympathetic neuronal pathways (18), and recordings can readily be made from these systems. Viral transsynaptic tracing has also been used extensively to identify anatomical circuits controlling visceral function (22, 34) and can identify multisynaptic connections with specific brain regions (3, 4). Combining EGFP identification with viral transneuronal labeling of multisynaptic pathways, as demonstrated in this study, is a powerful and potentially important technique for examining synaptic electrophysiology in functionally identified neural circuits.
Acknowledgments
We thank Christine Tomlinson for excellent technical assistance and Dr. Malcolm Ogilvie for assistance in preparing the figures. This work was supported by National Institutes of Health Grants MH 59995 (F.E.D.), NS 33506 (L.W.E.), NS 35366 and NS 35615 (G.E.P.), a postdoctoral fellowship from the Medical Research Council of Canada (B.W.B.), and a grant from the U.S. Department of Agriculture (C.L.W.).
Abbreviations
- PRV
pseudorabies virus
- EGFP
enhanced green fluorescent protein
- SCN
suprachiasmatic nucleus
- IGL
intergeniculate leaflet
- RGC
retinal ganglion cell
- EPSC
excitatory postsynaptic current
- IPSC
inhibitory postsynaptic current
- CNS
central nervous system
- PT
pretectum
References
- 1.Card J P. Anat Rec. 1998;253:176–185. doi: 10.1002/(SICI)1097-0185(199812)253:6<176::AID-AR6>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 2.Card J P, Enquist L W, Moore R Y. J Comp Neurol. 1999;407:438–452. doi: 10.1002/(sici)1096-9861(19990510)407:3<438::aid-cne11>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 3.Chen S, Yang M, Miselis R R, Aston-Jones G. Brain Res. 1999;838:171–183. doi: 10.1016/s0006-8993(99)01680-7. [DOI] [PubMed] [Google Scholar]
- 4.Enquist L W, Husak P J, Banfield B W, Smith G A. Adv Virus Res. 1999;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 5.Kuypers H G J M, Ugolini G. Trends Neurosci. 1990;13:71–75. doi: 10.1016/0166-2236(90)90071-h. [DOI] [PubMed] [Google Scholar]
- 6.Card J P. Neurosci Biobehav Rev. 1998;22:685–694. doi: 10.1016/s0149-7634(98)00007-4. [DOI] [PubMed] [Google Scholar]
- 7.Card J P, Whealy M E, Robbins A K, Moore R Y, Enquist L W. Neuron. 1991;6:957–969. doi: 10.1016/0896-6273(91)90236-s. [DOI] [PubMed] [Google Scholar]
- 8.Moore R Y, Speh J C, Card J P. J Comp Neurol. 1995;352:351–366. doi: 10.1002/cne.903520304. [DOI] [PubMed] [Google Scholar]
- 9.Provencio I, Cooper H M, Foster R G. J Comp Neurol. 1998;395:417–439. doi: 10.1002/(sici)1096-9861(19980615)395:4<417::aid-cne1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 10.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H J, Enquist L W. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiraly M, Dolivo M. Brain Res. 1982;240:43–54. doi: 10.1016/0006-8993(82)90642-4. [DOI] [PubMed] [Google Scholar]
- 12.Mayer M L, James M H, Russel R J, Kelly J S, Pasternak C A. J Neurosci. 1986;6:391–402. doi: 10.1523/JNEUROSCI.06-02-00391.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz L C, Burkhalter A, Dreyer W J. Nature (London) 1984;310:498–500. doi: 10.1038/310498a0. [DOI] [PubMed] [Google Scholar]
- 14.Ramoa A S, Campbell G, Shatz C J. Science. 1987;237:522–525. doi: 10.1126/science.3603038. [DOI] [PubMed] [Google Scholar]
- 15.Viana F, Gibbs L, Berger A J. Neuroscience. 1990;38:829–841. doi: 10.1016/0306-4522(90)90075-f. [DOI] [PubMed] [Google Scholar]
- 16.Spergel D J, Kruth U, Hanley D F, Sprengel R, Seeburg P H. J Neurosci. 1999;19:2037–2050. doi: 10.1523/JNEUROSCI.19-06-02037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickard G E, Smith B N, Belenky M, Rea M A, Dudek F E, Sollars P J. J Neurosci. 1999;19:4034–4045. doi: 10.1523/JNEUROSCI.19-10-04034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strack A M, Lowey A D. J Neurosci. 1990;10:2139–2147. doi: 10.1523/JNEUROSCI.10-07-02139.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strecker G J, Wuarin J P, Dudek F E. J Neurophysiol. 1997;78:2217–2220. doi: 10.1152/jn.1997.78.4.2217. [DOI] [PubMed] [Google Scholar]
- 20.Card J P, Rinaman L, Lynn R B, Lee B, Meade R P, Miselis R R, Enquist L W. J Neurosci. 1993;13:2515–2539. doi: 10.1523/JNEUROSCI.13-06-02515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jons A, Mettenleiter T C. J Virol Methods. 1997;66:283–292. doi: 10.1016/s0166-0934(97)00065-7. [DOI] [PubMed] [Google Scholar]
- 22.Ueyama T, Krout K E, Nguyen X V, Karpitskiy V, Kollert A, Mettenleiter T C, Loewy A D. Nat Neurosci. 1999;2:1051–1053. doi: 10.1038/15973. [DOI] [PubMed] [Google Scholar]
- 23.Pickard G E, Silverman A J. J Comp Neurol. 1981;196:155–172. doi: 10.1002/cne.901960111. [DOI] [PubMed] [Google Scholar]
- 24.Tian N, Hwang T N, Copenhagen D R. J Neurophysiol. 1998;80:1327–1340. doi: 10.1152/jn.1998.80.3.1327. [DOI] [PubMed] [Google Scholar]
- 25.Rorig B, Grantyn R. Dev Brain Res. 1993;74:98–110. doi: 10.1016/0165-3806(93)90088-r. [DOI] [PubMed] [Google Scholar]
- 26.Gao F, Wu S M. J Neurophysiol. 1998;80:1752–1764. doi: 10.1152/jn.1998.80.4.1752. [DOI] [PubMed] [Google Scholar]
- 27.Protti D A, Gerschenfeld H M, Llano I. J Neurosci. 1997;17:6075–6085. doi: 10.1523/JNEUROSCI.17-16-06075.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sterling P. In: The Synaptic Organization of the Brain. Shepard G M, editor. New York: Oxford Univ. Press; 1998. pp. 205–253. [Google Scholar]
- 29.Saito H A. Vision Res. 1983;23:1299–1308. doi: 10.1016/0042-6989(83)90105-0. [DOI] [PubMed] [Google Scholar]
- 30.Owczarzak M T, Pourcho R G. Anat Rec. 1999;255:363–373. doi: 10.1002/(SICI)1097-0185(19990801)255:4<363::AID-AR1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Pu M. J Comp Neurol. 1999;414:267–274. [PubMed] [Google Scholar]
- 32.Clarke R J, Ikeda H. Exp Brain Res. 1985;57:224–232. doi: 10.1007/BF00236527. [DOI] [PubMed] [Google Scholar]
- 33.Meijer J H, Rietveld W J. Physiol Rev. 1989;69:671–707. doi: 10.1152/physrev.1989.69.3.671. [DOI] [PubMed] [Google Scholar]
- 34.Card J P, Rinaman L, Schwaber J S, Miselis R R, Whealy M E, Robbins A K, Enquist L W. J Neurosci. 1990;10:1974–1994. doi: 10.1523/JNEUROSCI.10-06-01974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]