Abstract
The properties of the distribution of deleterious mutational effects on fitness (DDME) are of fundamental importance for evolutionary genetics. Since it is extremely difficult to determine the nature of this distribution, several methods using various assumptions about the DDME have been developed, for the purpose of parameter estimation. We apply a newly developed method to DNA sequence polymorphism data from two Drosophila species and compare estimates of the parameters of the distribution of the heterozygous fitness effects of amino acid mutations for several different distribution functions. The results exclude normal and gamma distributions, since these predict too few effectively lethal mutations and power-law distributions as a result of predicting too many lethals. Only the lognormal distribution appears to fit both the diversity data and the frequency of lethals. This DDME arises naturally in complex systems when independent factors contribute multiplicatively to an increase in fitness-reducing damage. Several important parameters, such as the fraction of effectively neutral non-synonymous mutations and the harmonic mean of non-neutral selection coefficients, are robust to the form of the DDME. Our results suggest that the majority of non-synonymous mutations in Drosophila are under effective purifying selection.
Keywords: Drosophila, distribution of mutational effects, lethals, lognormal, gamma, power law
1. Introduction
Recent advances in evolutionary genetics have led to a number of approaches for estimating the distribution of deleterious mutational effects on fitness (DDME) of non-synonymous mutations, using data on between-species sequence divergence and/or within-species sequence diversity (Bustamante et al. 2003; Nielsen & Yang 2003; Piganeau & Eyre-Walker 2003; Sawyer et al. 2003; Loewe et al. 2006). All assume a specific type of distribution of selection coefficients, which is then used to fit the data. Previous investigations have used a variety of distributions, including the normal, exponential and gamma distributions (Bustamante et al. 2003; Nielsen & Yang 2003; Piganeau & Eyre-Walker 2003; Sawyer et al. 2003; Loewe et al. 2006). The latter is widely used because of its convenient two-parameter form, which allows a wide range of curve shapes. However, none of these distributions has a special status, since there is currently no basis for a rational choice.
Here, we propose rejection of a candidate DDME if it cannot explain (i) DNA sequence diversity data in two related species with different effective population sizes, and (ii) the frequency of dominant, effectively lethal mutations caused by amino acid mutations. We find that a lognormal DDME satisfies these conditions much better than a gamma distribution or a power law.
2. Material and methods
We define the DDME as the genome-wide distribution of the heterozygous selection coefficient, s, associated with a new deleterious, non-synonymous mutation. We use diversity data from 17 loci of Drosophila miranda and 14 loci of Drosophila pseudoobscura, two closely related species of fruitfly with similar habitats, but significantly different effective population sizes (Ne) as estimated from their silent nucleotide site diversities, πS (Loewe et al. 2006). The similarity of the two species means that they probably share the same DDME, so that the larger Ne of D. pseudoobscura compared with D. miranda causes a larger fraction of sites to experience effective purifying selection. This results in a smaller increase in non-synonymous diversity, πA, than in πS. Assuming a given type of DDME for mutations affecting amino acid sites and a fraction of completely neutral, non-synonymous mutations (cn), we can calculate the expectation of πA for each species. By equating these to the pair of observed mean values of πA, we estimate the parameters of the DDME, assuming that it can be described by two parameters. Our method assumes approximate mutation–selection-drift equilibrium and independence among non-synonymous polymorphisms, but is not affected by the details of the frequency distributions of variants. It should, therefore, provide robust estimates of the parameters of the DDME for a fixed value of cn. Statistical accuracy is assessed by computing 1000 bootstraps. To improve analysis, we used the diversity index, DI (the ratio of the values πA/πS for the two species) to eliminate the 12.2% of all bootstraps with DI≤1, since these cannot be explained by any plausible model. Further details are described in the electronic supplementary material and by Loewe et al. (2006).
3. Results
To test whether a DDME that is compatible with observed diversity data also satisfies our second criterion requires an estimate of the rate at which non-synonymous mutations with effectively lethal (i.e. lethal or sterile) heterozygous effects arise. The difficulty is that most lethal mutations are recessive and it is hard to study those that are not. While point mutations, indels and transposable elements (TEs) can all induce dominant, effectively lethal mutations, we are only concerned with non-synonymous mutations. It is virtually impossible to estimate the rate of spontaneous dominant lethal mutations, and most of these are probably due to chromosome breaks (Ashburner 1989). However, the results of ethylmethane sulphonate (EMS) mutagenesis, which mainly but not exclusively induces point mutations, suggest that dominant female sterile mutations arise in D. melanogaster at about 1/500th of the rate for recessive lethal mutations (Ashburner 1989). Molecular analyses of two of the genes concerned show that the majority of the mutational lesions involved are non-synonymous mutations (Timinszky et al. 2002; Venkei & Szabad 2005).
The approximate overall frequency of such mutations can be assessed as follows. Recent data suggest that spontaneous recessive lethal mutations arise at a rate of about 0.045 per zygote per generation in D. melanogaster, but many of these are probably due to TE insertions (Charlesworth et al. 2004). The rate of TE insertions is about 0.2 per zygote per generation (Maside et al. 2000). Assuming that 25% of the genome is coding sequence (Misra et al. 2002) and 25% of all genes are vital (Oh et al. 2003) gives an estimate of 0.0125 recessive lethal TE-insertions per zygote per generation. If 25% of non-TE mutations are indels (Charlesworth et al. 2004), this suggests that the recessive lethal mutation rate due to base substitutions is around 0.75×(0.045−0.0125)=0.024. The rate of mutation to dominant female sterile non-synonymous mutations in Drosophila is thus about 5×10−5. If dominant lethal and dominant male sterility mutations arise at a similar rate, the net rate of mutation to effectively lethal dominant mutations is about 2×10−4. There is 27.8 Mb of exon sequence in D. melanogaster (Misra et al. 2002); about 70% of these sites can generate non-synonymous mutations without shifting the reading frame. With a mutation rate of 1.5×10−9 (Loewe et al. 2006), this results in a total mutation rate for non-synonymous mutations of U=0.058 per zygote per generation. The fraction λ of non-synonymous mutations that are dominant effective lethals is thus about 3.4×10−3. The precise value of this parameter is not important; however, the fact that effectively lethal non-synonymous mutations occur at a detectable, but low rate means that any candidate DDME that predicts either no or many such mutations can be rejected.
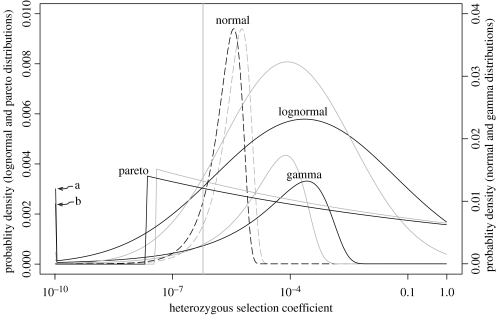
Table 1 gives an overview of the various types of DDME that were tested against the data. Figure 1 plots the best-fitting estimates for visual inspection. Table 2 reports the distributional parameters, as well as scaled measures of selection intensities, for the DDMEs that can be fitted to the diversity data (which is not the case for the normal distribution with cn=0 or 2.5%). The gamma distribution fits the diversity data well (Loewe et al. 2006). The number of dominant effective lethals predicted by the best gamma DDME is, however, very small, because the diversity data can only be fitted by a gamma distribution with a relatively small width (table 2), so that the right-hand end of the distribution falls off quickly (figure 1). Inspection of the results for the gamma distribution shows that 6.5% of the 802 bootstraps that could be fitted to the data for cn=0 lead to potentially realistic genomic lethal rates (Uλ between 10−5 and 0.004), but most results gave unacceptably low values. Thus, the gamma DDME does not easily predict plausible numbers of these mutations.
Table 1.
Candidate distributions of mutational effects.
| DDME | probability density function | intuitive meaning |
|---|---|---|
| normal | Arises from the central limit theorem if a mutation has effects on several different traits that affect fitness additively. Determined by location parameter μ (mean) and shape parameter σ (s.d.). | |
| gamma | ϕ(s)=(sα−1e−s/β)/(βαΓ(α)) | Arises naturally as time-to-first-fail for a system with α−1 standby backups that have exponentially distributed life times with parameter β (NIST/SEMATECH 2005), where α is the shape and β is the location. |
| pareto | Arises if a stochastic process that is expected to grow exponentially is killed (or observed) randomly (Reed & Hughes 2002). Determined by location smin (=smallest value) and shape k. | |
| lognormal | Arises naturally from the central limit theorem if many small pleiotropic fitness-reducing effects have a multiplicative, cumulative effect on damage (NIST/SEMATECH 2005). We use location parameter μg (=geometric mean=median=eμ) and shape parameter σg (=eσ). |
Figure 1.
Probability densities fitted to the diversity data for various distributions of mutational effects (DDMEs). Solid lines indicate cases where the data could be fitted; dashed lines could not be fitted. Black lines assume cn=0, grey lines cn=2.5%. The scales for the lognormal and pareto distributions are on the left-hand y-axis and those for the normal and gamma distributions are on the right-hand y-axis. The vertical grey line denotes Nes=0.5 for D. miranda. The spikes at 10−10 integrate over all probability mass down to 0, where a is for the lognormal (cn=0) and b is for the gamma (cn=0). The normal DDMEs shown give the closest fit to the data that we could find for the given cn values. They consist of 48% advantageous mutations (truncated). Fine-tuning DDME location and cn allows fits of many DDMEs (including constant s) to diversity data, but not to the frequency of lethals.
Table 2.
Numerical details of the distributions of deleterious mutational effects. All estimates in this table are based on diversity data for D. miranda (mir, Ne=837 000) and D. pseudoobscura (pso, Ne=4 770 000; Loewe et al. 2006). Numerical results assume a mutation rate of u=1.5×10−9, a mutational bias of κ=2 (the ratio of mutation rates to and from deleterious amino acids) and Nes=0.5 as the border to effective neutrality (Loewe et al. 2006). Values in parentheses give the lower 5th percentiles and the upper 5th percentiles from 1000 bootstraps. The different columns have the following meanings. DDME, cn%: type of distribution and the fraction of completely neutral non-synonymous mutations assumed. Shape, location: the parameters that determine the distributions (see table 1). Species: the focal species whose Ne was used to compute the next six columns. Nes (am), (hm): the arithmetic and harmonic mean, respectively, of all effectively deleterious, but non-lethal selection coefficients, multiplied by effective population size. These values (and those in the next three columns) use all s values that are larger than the border to effective neutrality and smaller than lethal (s=1). Nes (5%), (95%): the lower and upper 5% percentiles of the truncated DDME. CV: coefficient of variation of the truncated DDME. cne (%): the fraction of effectively neutral non-synonymous mutations. λ(%): fraction of the DDME with s≥1 (effectively dominant lethals).
| DDME cn (%) | shape | location | species | Nes (am) | Nes (hm) | Nes (5%) | Nes (95%) | CV | cne (%) | λ (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| log N | 202 | 0.000231 | mir | 29900 | 8.47 | 1.21 | 190000 | 3.34 | 13.2 | 5.73 |
| 0% | (9.34/2490) | (8.29×10−6/0.00448) | (108/56600) | (3.65/10.7) | (0.781/1.54) | (362/398000) | (2.45/12.3) | (8.62/17.4) | (9×10−6/23.8) | |
| pso | 159000 | 12.6 | 1.99 | 999000 | 3.47 | 7.44 | ||||
| (546/309000) | (7.80/16.2) | (1.31/2.73) | (1810/2190000) | (2.53/12.9) | (1.73/11.0) | |||||
| log N | 44.9 | 8.41×10−5 | mir | 9880 | 7.44 | 1.19 | 35100 | 5.26 | 12.1 | 0.681 |
| 2.5% | (4.14/1480) | (5.90×10−6/0.00421) | (16.6/53700) | (3.22/11.5) | (0.801/1.71) | (62.4/370000) | (2.26/11.5) | (6.03/16.7) | (2×10−15/22.4) | |
| pso | 52900 | 13.4 | 2.31 | 184000 | 5.44 | 6.43 | ||||
| (85.9/306000) | (8.69/21.1) | (1.49/4.54) | (328/2130000) | (2.33/11.9) | (2.64/11.3) | |||||
| gamma | 0.299 | 0.00089 | mir | 255 | 8.35 | 1.31 | 1100 | 1.67 | 12.7 | <10−100 |
| 0% | (0.0782/0.741) | (1.60×10−5/209000) | (11.6/95700) | (3.49/17.9) | (0.843/3.01) | (36.9/558000) | (1.08/2.15) | (7.91/17.0) | (<10−100/61) | |
| pso | 1370 | 14.4 | 2.5 | 6220 | 1.74 | 7.57 | ||||
| (55.3/521000) | (8.69/25.0) | (1.55/5.47) | (188/3110000) | (1.14/2.22) | (2.35/13.4) | |||||
| gamma | 0.448 | 0.000171 | mir | 70.6 | 6.86 | 1.21 | 274 | 1.39 | 11.4 | <10−100 |
| 2.5% | (0.0996/1.32) | (5.45×10−6/956) | (6.96/76500) | (3.19/17.2) | (0.877/2.96) | (18.4/463000) | (0.832/2.08) | (5.73/16.6) | (<10−100/45.2) | |
| pso | 382 | 14.7 | 2.73 | 1550 | 1.45 | 6.58 | ||||
| (35.4/392000) | (9.48/28.4) | (1.77/7.31) | (97.2/2450000) | (0.865/2.13) | (2.87/13.4) | |||||
| pareto | 0.0458 | 2.12×10−8 | mir | 43400 | 5.54 | 0.823 | 323000 | 2.9 | 14.3 | 44.5 |
| 0% | (0.0154/0.101) | (5.61×10−11/1.09×10−7) | (27500/58700) | (4.21/6.47) | (0.72/0.938) | (183000/422000) | (2.59/3.58) | (11.0/19.4) | (19.5/69.9) | |
| pso | 210000 | 5.82 | 0.861 | 1490000 | 3.18 | 7.21 | ||||
| (124000/274000) | (4.65/7.20) | (0.759/1.01) | (766000/2020000) | (2.80/3.97) | (0.454/13.4) | |||||
| pareto | 0.0494 | 3.71×10−8 | mir | 42300 | 5.43 | 0.819 | 304000 | 2.94 | 15.2 | 43 |
| 2.5% | (0.0148/0.114) | (7.43×10−11/1.60×10−7) | (25100/57900) | (3.93/6.48) | (0.708/0.937) | (157000/421000) | (2.59/3.79) | (12.4/19.6) | (16.3/70.8) | |
| pso | 204000 | 5.7 | 0.857 | 1480000 | 3.22 | 7.53 | ||||
| (114000/275000) | (4.80/7.52) | (0.769/1.23) | (668000/2030000) | (2.80/4.18) | (2.58/14.1) |
Recent searches for general principles have frequently uncovered or attempted to fit power laws (Reed & Hughes 2002; Mitzenmacher 2004), and so a power law such as the pareto distribution (table 1) might be a good candidate DDME. While we found that a pareto DDME can fit the diversity data reasonably well, it failed to predict plausible fractions of lethals (table 2). In contrast, a lognormal DDME can fit both observed diversities and fractions of lethals. This result seems to be relatively insensitive to different assumptions about cn, and the bootstraps for Uλ largely overlap the range of values that are consistent with the data. For a lognormal DDME and cn=0, a much larger fraction of bootstraps (60% out of 610) predicts a plausible genomic lethal rate than with a gamma DDME. The fact that this fraction is not higher is probably due to the limited size of our dataset and the correspondingly noisy statistics.
4. Discussion
Is there a theoretical reason for the relatively good performance of the lognormal distribution? The central limit theorem, which states that a variable affected by independent additive effects of several other variables is normally distributed, implies that a variable controlled multiplicatively by several independent factors follows the lognormal distribution (Koch 1969; Mitzenmacher 2004; NIST/SEMATECH 2005). Darwinian fitness must be affected by many different factors operating at different levels of biological organization. We suggest that the extent of a reduction in fitness caused by a deleterious mutation, as measured by s, is a function of the amount of damage that it causes at several independent functional levels. If the total amount of damage were a multiplicative function of the amounts of damage at each level, a lognormal distribution of s would result (NIST/SEMATECH 2005).
Regardless of the question of the true nature of the DDME, the results presented in table 2 are encouraging in that some of the more important parameters derived from its properties are relatively invariant with respect to the type of distribution and are also fairly well bounded by the bootstrap procedure. In particular, the harmonic mean of Nes for effectively deleterious mutations is fairly similar for the different distributions and its bootstrap intervals are bounded well above 1. As noted previously, this parameter is close to the mean selection coefficient associated with polymorphic mutations that are not effectively neutral i.e. have Nes>0.5 (Loewe et al. 2006). It plays an important role in processes such as background selection and Muller's ratchet (Charlesworth & Charlesworth 2000). Similarly, the proportion of effectively neutral mutations is consistently estimated to be less than 20% and usually less than 10% (table 2). These conclusions are consistent with results from other methods (Bustamante et al. 2003; Nielsen & Yang 2003; Piganeau & Eyre-Walker 2003; Sawyer et al. 2003; Loewe et al. 2006).
It is likely that datasets of larger diversity and more accurate estimates of the fraction of dominant effective lethals will lead to more precise estimates in the future. Obviously, it is possible that there could be a mixture of distributions, with widely different means, contributing to the overall DDME and mimicking the results we have obtained by fitting the lognormal. For practical purposes, it is preferable to use a single distribution that matches the data successfully.
The results strengthen the conclusion that most amino acid mutations segregating in natural populations of Drosophila have sufficiently large deleterious effects on fitness that they behave quasi-deterministically (Nes>1), although large Ne values imply that selection against deleterious mutations in Drosophila is mostly weak (Loewe et al. 2006; mean s≈10−4). Despite the very small selection coefficients associated with most mutations, our best estimates for the lognormal DDME predict appreciable numbers of mutations with detectable fitness effects; we expect 11 or 7% of all non-synonymous mutations to have heterozygous effects between s=0.01−0.1, assuming cn=0% or 2.5%, respectively. Recent experiments on isolating EMS mutations in specific Drosophila genes by TILLING have shown that a substantial fraction of non-synonymous mutations can have drastic homozygous fitness effects (Winkler et al. 2005). Together with earlier findings that EMS-induced mutations with drastic homozygous fitness effects typically have small, but detectable fitness losses when heterozygous (Simmons et al. 1978), this is consistent with the predictions of the lognormal DDME, as is the fact that many human dominant Mendelian disorders are caused by single amino acid changes (Yampolsky et al. 2005). The hypothesis that there is a substantial minority of amino acid mutations with experimentally detectable heterozygous fitness effects can be tested in model organisms such as yeast or Drosophila, by measuring the fitness effects of induced non-synonymous mutations of known identity.
Acknowledgments
We thank Jay Taylor for helpful discussions and the Royal Society and the Leverhulme foundation for support. We have no competing financial interests.
Supplementary Material
References
- Ashburner M. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. Drosophila: a laboratory handbook. [Google Scholar]
- Bustamante C.D, Nielsen R, Hartl D.L. Maximum likelihood and Bayesian methods for estimating the distribution of selective effects among classes of mutations using DNA polymorphism data. Theor. Popul. Biol. 2003;63:91–103. doi: 10.1016/s0040-5809(02)00050-3. doi:10.1016/S0040-5809(02)00050-3 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Phil. Trans. R. Soc. B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. doi:10.1098/rstb.2000.0717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Borthwick H, Bartolome C, Pignatelli P. Estimates of the genomic mutation rate for detrimental alleles in Drosophila melanogaster. Genetics. 2004;167:815–826. doi: 10.1534/genetics.103.025262. doi:10.1534/genetics.103.025262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A.L. The logarithm in biology II. Distributions simulating the log-normal. J. Theor. Biol. 1969;23:251–268. doi: 10.1016/0022-5193(69)90040-x. doi:10.1016/0022-5193(69)90040-X [DOI] [PubMed] [Google Scholar]
- Loewe L, Charlesworth B, Bartolomé C, Nöel V. Estimating selection on non-synonymous mutations. Genetics. 2006;172:1079–1092. doi: 10.1534/genetics.105.047217. doi:10.1534/genetics.105.047217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maside X, Assimacopoulos S, Charlesworth B. Rates of movement of transposable elements on the second chromosome of Drosophila melanogaster. Genet. Res. 2000;75:275–284. doi: 10.1017/s0016672399004474. doi:10.1017/S0016672399004474 [DOI] [PubMed] [Google Scholar]
- Misra S, et al. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0083. doi:10.1186/gb-2002-3-12-research0083 research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitzenmacher M. A brief history of generative models for power law and lognormal distributions. Internet Math. 2004;1:226–251. [Google Scholar]
- Nielsen R, Yang Z. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 2003;20:1231–1239. doi: 10.1093/molbev/msg147. doi:10.1093/molbev/msg147 [DOI] [PubMed] [Google Scholar]
- NIST/SEMATECH 2005 8.1.6 What are the basic lifetime distribution models used for non-repairable populations? In e-handbook of statistical methods, see http://www.itl.nist.gov/div898/handbook/apr/section1/apr16.htm
- Oh S.W, et al. A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics. 2003;163:195–201. doi: 10.1093/genetics/163.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau G, Eyre-Walker A. Estimating the distribution of fitness effects from DNA sequence data: implications for the molecular clock. Proc. Natl Acad. Sci. USA. 2003;100:10 335–10 340. doi: 10.1073/pnas.1833064100. doi:10.1073/pnas.1833064100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed W.J, Hughes B.D. From gene families and genera to incomes and internet file sizes: why power laws are so common in nature. Phys. Rev. E. 2002;66 doi: 10.1103/PhysRevE.66.067103. doi:10.1103/PhysRevE.66.067103 art. no.-067103. [DOI] [PubMed] [Google Scholar]
- Sawyer S.A, Kulathinal R.J, Bustamante C.D, Hartl D.L. Bayesian analysis suggests that most amino acid replacements in Drosophila are driven by positive selection. J. Mol. Evol. 2003;57:S154–S164. doi: 10.1007/s00239-003-0022-3. doi:10.1007/s00239-003-0022-3 [DOI] [PubMed] [Google Scholar]
- Simmons M.J, Sheldon E.W, Crow J.F. Heterozygous effects on fitness of EMS-treated chromosomes in Drosophila melanogaster. Genetics. 1978;88:575–590. doi: 10.1093/genetics/88.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G, et al. The importin-beta P446L dominant-negative mutant protein loses RanGTP binding ability and blocks the formation of intact nuclear envelope. J. Cell Sci. 2002;115:1675–1687. doi: 10.1242/jcs.115.8.1675. [DOI] [PubMed] [Google Scholar]
- Venkei Z, Szabad J. The KavarD dominant female-sterile mutations of Drosophila reveal a role for the maternally provided α-tubulin4 isoform in cleavage spindle maintenance and elongation. Mol. Genet. Genomics. 2005;273:283–289. doi: 10.1007/s00438-005-1109-x. doi:10.1007/s00438-005-1109-x [DOI] [PubMed] [Google Scholar]
- Winkler S, et al. Target-selected mutant screen by TILLING in Drosophila. Genome Res. 2005;15:718–723. doi: 10.1101/gr.3721805. doi:10.1101/gr.3721805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yampolsky L.Y, Kondrashov F.A, Kondrashov A.S. Distribution of the strength of selection against amino acid replacements in human proteins. Hum. Mol. Genet. 2005;14:3191–3201. doi: 10.1093/hmg/ddi350. doi:10.1093/hmg/ddi350 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.