Abstract
Amphibians tend to exhibit conservative morphological evolution, and the application of molecular and bioacoustic tools in systematic studies have been effective at revealing morphologically ‘cryptic’ species within taxa that were previously considered to be a single species. We report molecular genetic findings on two forest-dwelling ranid frogs from localities across Southeast Asia, and show that sympatric evolutionary lineages of morphologically cryptic frogs are a common pattern. These findings imply that species diversity of Southeast Asian frogs remains significantly underestimated, and taken in concert with other molecular investigations, suggest there may not be any geographically widespread, forest-dwelling frog species in the region. Accurate assessments of diversity and distributions are needed to mitigate extinctions of evolutionary lineages in these threatened vertebrates.
Keywords: biodiversity, speciation, conservation, amphibians
1. Introduction
Despite a worldwide decline in amphibian populations (Stuart et al. 2004), the number of recognized species of amphibians has increased dramatically in recent years (Glaw & Köhler 1998; Hanken 1999; Meegaskumbura et al. 2002; Köhler et al. 2005; Ron et al. 2006), making amphibians one of the vertebrate groups with the highest proportional rate of description of new species (Hanken 1999). Amphibians tend to exhibit conservative morphological evolution (Cherry et al. 1978), and the application of molecular genetic and bioacoustic tools in systematic studies have been particularly effective at revealing morphologically ‘cryptic’ species within taxa that were previously considered to be a single species (e.g. Bogart & Tandy 1976; Hillis et al. 1983; Highton 1989; Wynn & Heyer 2001; Gower et al. 2005).
We examined genetic variation across the geographic ranges of two Southeast Asian ranid frogs. Odorrana livida and Rana chalconota live in intact forest, along cascading streams (livida) or slow-moving streams and swamps (chalconota). Both frogs have been hypothesized to represent a complex of species across their ranges, but identifications of members of these complexes have been greatly confused due to their morphological similarity (Iskandar & Colijn 2000; Bain et al. 2003). Odorrana livida was originally described from Myanmar and has been reported from India to Vietnam (Bain et al. 2003). Rana chalconota was originally described from Java and has been reported from peninsular Thailand, peninsular Malaysia, Sumatra and Borneo (Boulenger 1920; Iskandar & Colijn 2000).
2. Material and methods
We sampled frogs from across Southeast Asia that were identified in taxonomic studies on the basis of morphology as O. livida and R. chalconota (Boulenger 1920; Taylor 1962; Inger 1966; Inger & Chanard 1997; Bain et al. 2003). We sequenced and analysed 2150 aligned characters of mitochondrial (mt) DNA (partial COXIII, complete tRNAGly, complete ND3, partial tRNAArg, partial 16S, partial tRNAMet, complete ND2, and partial tRNATrp genes) from frogs morphologically resembling O. livida and 1082 aligned characters of mt DNA (partial COXIII, complete tRNAGly, complete ND3, partial tRNAArg, partial 16S genes) from frogs morphologically resembling R. chalconota. We also used historic DNA methods to obtain 600 and 361 nucleotide base pairs (bp) of the 16S gene, respectively, from the neotype museum specimen of O. livida (collected in 1887) and the syntype museum specimen of its junior synonym O. chloronota (collected prior to 1875). See electronic supplementary material for voucher information, sequencing protocols, GenBank accession numbers and methods of phylogenetic analyses.
3. Results
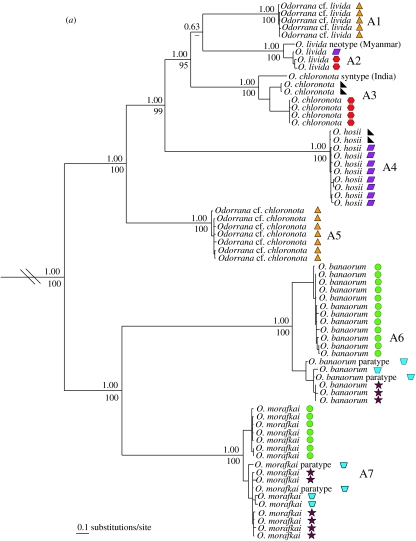
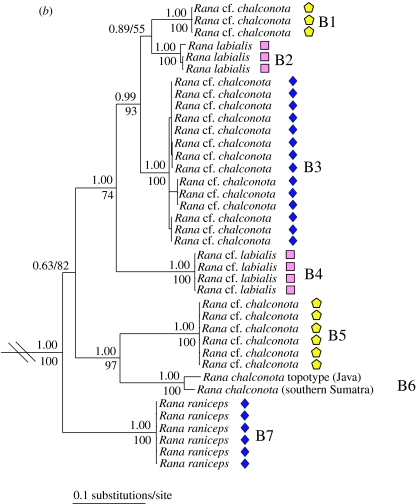
Mitochondrial DNA sequences from both frog species complexes unexpectedly reveal two deeply divergent lineages occurring in sympatry (uncorrected pairwise sequence divergences of 9.95–16.12% between sympatric lineages of O. livida and 10.79–15.21% between sympatric lineages of R. chalconota) at 10 localities (figure 1), and phylogenetic analyses show that sympatric lineages are usually not each other's closest relatives (figure 2). Sorting voucher specimens a posteriori by mt DNA lineage illuminates at least one diagnostic morphological character (in body size, coloration in life, pattern on the back and rear of the thigh, presence/absence of spinules on the body, presence/absence of gular pouches and condition of the nuptial pads) in all but two lineages (the allopatric lineages A3 and A5; figure 2). We hypothesize that these mt DNA lineages represent distinct species on the basis of their sympatric occurrences and long evolutionary history of isolation, as inferred by deep genetic divergences and, in most cases, diagnostic morphological characters. Names can be assigned to eight of the 14 recovered clades by resurrecting old, junior synonyms (A3, B2, B7; figure 2), correctly applying commonly used names (A2, A4, B6; figure 2), and recognizing newly described species (A6–A7; figure 2), while the remaining six clades represent species undescribed to science (A1, A5, B1, B3, B4–B5; figure 2). True O. livida and R. chalconota were identified from an historical DNA sequence obtained from a type museum specimen preserved since 1887 (A2; figure 2) or from recent material collected at the type locality (B6; figure 2), and these two species actually occupy only small portions of their published geographic ranges.
Figure 1.
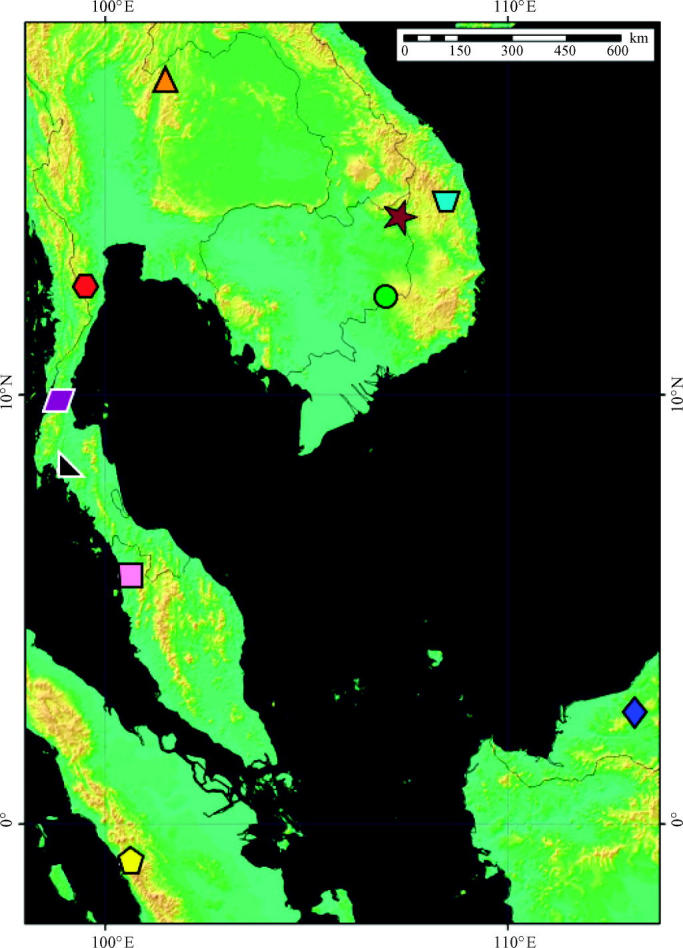
Map illustrating localities where two sympatric, morphologically ‘cryptic’ lineages of frogs were sampled in this study. Localities are Phu Luang, Loei Prov., Thailand (equilateral triangle); Khao Phanom Bencha, Krabi Prov., Thailand (right-angle triangle); Kaeng Krachan, Prachuap Kirikhan Prov., Thailand (hexagon); Namtok Ngao, Ranong Prov., Thailand (parallelogram); O'Rang, Mondolkiri Prov., Cambodia (circle); Ta Veng, Ratanakiri Prov., Cambodia (star); An Khe, Gia Lai Prov., Vietnam (trapezoid); Padang, West Sumatra (pentagon); Gunong Jerai, Peninsular Malaysia (square); Bukit Sarang, Sarawak (diamond).
Figure 2.
Fifty percent majority-rule consensus phylograms resulting from mixed-model Bayesian analyses of mitochondrial DNA from frogs morphologically resembling (a) Odorrana livida and (b) Rana chalconota. Trees were rooted with (a) O. cf. chapaensis and O. bacboensis and (b) R. cubitalis and R. erythraea. Numbers above and below nodes are Bayesian posterior probabilities and parsimony bootstrap values greater than 50, respectively. Maximum parsimony analyses recovered the same topologies except that O. chloronota and O. livida were hypothesized to be sister clades (bootstrap 58%). Symbols refer to localities shown in figure 1.
4. Discussion
Our findings of multiple cases of morphologically cryptic species in sympatry across the geographic ranges of two frog species complexes imply that amphibian species diversity remains significantly underestimated in Southeast Asia. Frogs that have been identified on the basis of morphology as O. livida and R. chalconota represent at least 14 species. We suggest that these species have been overlooked or confused due to their extreme morphological similarity compounded by an erroneous assumption that samples collected together are conspecific. Since sympatric lineages are usually not sister lineages, these frogs probably diversified by allopatric or parapatric speciation, with subsequent range shifts into sympatry. Many of the sympatric cryptic species were collected in syntopy, with individuals of different species found within centimetres of each other. It is not known how these morphologically similar species partition resources and maintain reproductive isolation, but investigations of microhabitat utilization (by adults and larvae) and vocalizations may offer insight into these topics.
To date, every molecular genetic study that has broadly sampled populations across the range of a widespread frog species in Southeast Asia (southern China to Sulawesi) has uncovered genetic diversity interpreted by those authors as unrecognized species diversity (Toda et al. 1998; Emerson et al. 2000; Li et al. 2001a,b; Matsui et al. 2001; Veith et al. 2001; Brown & Guttman 2002; Bain et al. 2003; Evans et al. 2003; Matsui et al. 2005). Seven of these ten studies involved frog species that live only in intact forest. Most of the new species identified by molecular approaches are allopatrically or parapatrically distributed, but cases of morphologically cryptic frog species occurring in sympatry have been revealed in Vietnam (Bain et al. 2003), Peninsular Malaysia (Narins et al. 1998) and Java (Toda et al. 1998; Veith et al. 2001). These studies and our results suggest there may not be any geographically widespread, forest-dwelling frog species in Southeast Asia, and that sympatric evolutionary lineages of morphologically cryptic frogs are a common pattern in the region.
The process of defining species boundaries is more than academic. Southeast Asia has the highest relative rate of deforestation of any major tropical region (Sodhi et al. 2004), and understanding which species occur where is essential to conservation managers mitigating loss of biodiversity. Single, widespread ‘species’ actually represent multiple species having smaller geographic ranges, and consequently, greater vulnerability to extinction. It is apparent that biodiversity inventories of frogs based on morphology alone can be misleading, and that the conservative morphological evolution of amphibians obscures divergent evolutionary lineages that warrant recognition and protection. Tissue sampling should become routine in amphibian inventories so that molecular genetic tools can play a significant role in more realistically defining amphibian species diversity in coming years.
Acknowledgments
We thank T. Chan-ard, Y. Chuaynkern, C. Chuechat, P. Francis, D. Gusman, D. Hon-Tjong, D. Iskandar, S. Makchai, T. Neang, J. Sheridan, K. Sok and F. Yulus for assistance with collecting specimens, and the National Research Council of Thailand, Royal Forest Department of Thailand, Thailand Natural History Museum and Wildlife Conservation Society for facilitating fieldwork. C. J. McCarthy, B. Clarke and M. Wilkinson (The Natural History Museum, London), R. W. Murphy (Royal Ontario Museum), T. Chan-ard, Y. Chuaynkern and J. Nabhitabhata (Thailand Natural History Museum), N. Yaakob and J. Sukumaran (Forest Research Institute Malaysia), D. R. Frost and L. Ford (American Museum of Natural History) and J. Campbell (University of Texas at Arlington) loaned specimens and tissues. R. Bain provided tissue from the BMNH type specimens with permission of C. J. McCarthy. K. Kline assisted with sequencing DNA. Sequencing was performed in The Field Museum's Pritzker Laboratory for Molecular Systematics and Evolution operated with support from the Pritzker Foundation. Bayesian analyses were executed on a computer cluster by R. Vogelbacher and the DePaul University Bioinformatics Group in conjunction with the Illinois Bio-Grid. The John D. and Catherine T. MacArthur Foundation and the Marshall Field III Fund at The Field Museum supported the research. This work was completed in partial fulfillment of the requirements for the doctoral degree (to B.L.S.) at the Graduate College of the University of Illinois at Chicago.
Supplementary Material
Voucher information, sequencing protocols, GenBank accession numbers and methods of phylogenetic analyses
References
- Bain R.H, Lathrop A, Murphy R.W, Orlov N.L, Ho C.T. Cryptic species of a cascade frog from Southeast Asia: taxonomic revisions and descriptions of six new species. Am. Mus. Novit. 2003;3417:1–60. doi:10.1206/0003-0082(2003)417<0001:CSOACF>2.0.CO;2 [Google Scholar]
- Bogart J.P, Tandy M. Polyploid amphibians: three more diploid–tetraploid cryptic species of frogs. Science. 1976;193:334–335. doi: 10.1126/science.935871. [DOI] [PubMed] [Google Scholar]
- Boulenger G.A. A monograph of the South Asian, Papuan, Melanesian and Australian frogs of the genus Rana. Rec. Indian Mus. 1920;20:1–226. [Google Scholar]
- Brown R.M, Guttman S.I. Phylogenetic systematics of the Rana signata complex of Philippine and Bornean stream frogs: reconsideration of Huxley's modification of Wallace's line at the Oriental–Australian faunal zone interface. Biol. J. Linn. Soc. 2002;76:393–461. doi:10.1046/j.1095-8312.2002.00062.x [Google Scholar]
- Cherry L.M, Case S.M, Wilson A.C. Frog perspective on the morphological difference between humans and chimpanzees. Science. 1978;200:209–211. doi: 10.1126/science.635583. [DOI] [PubMed] [Google Scholar]
- Emerson S.B, Inger R.F, Iskandar D. Molecular systematics and biogeography of the fanged frogs of Southeast Asia. Mol. Phylogenet. Evol. 2000;16:131–142. doi: 10.1006/mpev.2000.0778. doi:10.1006/mpev.2000.0778 [DOI] [PubMed] [Google Scholar]
- Evans B.J, Brown R.M, McGuire J.A, Supriatna J, Andayani N, Diesmos A, Iskandar D, Melnick D.J, Cannatella D.C. Phylogenetics of fanged frogs: testing biogeographical hypotheses at the interface of the Asian and Australian faunal zones. Syst. Biol. 2003;52:794–819. doi:10.1080/10635150390251063 [PubMed] [Google Scholar]
- Glaw F, Köhler J. Amphibian species diversity exceeds that of mammals. Herpetol. Rev. 1998;29:11–12. [Google Scholar]
- Gower D.J, Bahir M.M, Mapatuna Y, Pethiyagoda R, Raheem D, Wilkinson M. Molecular phylogenetics of Sri Lankan Ichthyophis (Amphibia: Gymnophiona: Ichthyophiidae), with discovery of a cryptic species. Raff. Bull. Zool. 2005;12:153–161. [Google Scholar]
- Hanken J. Why are there so many new amphibian species when amphibians are declining? Trends. Ecol. Evol. 1999;14:7–8. doi: 10.1016/s0169-5347(98)01534-1. doi:10.1016/S0169-5347(98)01534-1 [DOI] [PubMed] [Google Scholar]
- Highton R. Biochemical evolution in the slimy salamanders of the Plethodon glutinosus complex in the eastern United States. Part I. Geographic protein variation. Ill. Biol. Monog. 1989;57:1–78. [Google Scholar]
- Hillis D.M, Frost J.S, Wright D.A. Phylogeny and biogeography of the Rana pipiens complex: a biochemical evaluation. Syst. Zool. 1983;32:132–143. doi:10.2307/2413277 [Google Scholar]
- Inger R.F. The systematics and zoogeography of the Amphibia of Borneo. Fieldiana Zool. 1966;52:1–402. [Google Scholar]
- Inger R.F, Chanard T. A new species of ranid frog from Thailand, with comments on Rana livida (Blyth) Nat. Hist. Bull. Siam Soc. 1997;45:65–70. [Google Scholar]
- Iskandar D, Colijn E. Preliminary checklist of Southeast Asian and New Guinean herpetfauna [sic]. I. Amphibians. Treubia. 2000;31:1–133. [Google Scholar]
- Köhler J, Vieites D.R, Bonett R.M, Garciá F.H, Glaw F, Steinke D, Vences M. New amphibians and global conservation: a boost in species discoveries in a highly endangered vertebrate group. BioScience. 2005;55:693–696. [Google Scholar]
- Li C, Ye C.Y, Fei L. Taxonomic studies of Odorrana andersonii in China. Acta Zootaxon. Sin. 2001a;26:234–238. [Google Scholar]
- Li C, Ye C.-Y, Fei L. Taxonomic studies of Odorrana versabilis in China. I. Taxonomic status of the geographic populations. Acta Zootaxon. Sin. 2001b;26:593–600. [Google Scholar]
- Matsui M, Nishikawa K, Khonsue W, Panha S, Nabhitabhata J. Allozymic variation in Rana nigrovittata (Amphibia: Anura) within Thailand with special reference to the taxonomic status of R. mortenseni. Nat. Hist. J. Chulalongkorn Univ. 2001;1:15–22. [Google Scholar]
- Matsui M, Ito H, Shimada T, Ota H, Saidapur S.K, Khonsue W, Tanaka-Ueno T, Wu G.F. Taxonomic relationships within the pan-Oriental narrow-mouth toad Microhyla ornata as revealed by mtDNA analysis (Amphibia Anura, Microhylidae) Zool. Sci. 2005;22:489–495. doi: 10.2108/zsj.22.489. doi:10.2108/zsj.22.489 [DOI] [PubMed] [Google Scholar]
- Meegaskumbura M, Bossuyt F, Pethiyagoda R, Manamendra-Arachchi K, Bahir M, Milinkovitch M.C, Schneider C.J. Sri Lanka: an amphibian hot spot. Science. 2002;298:379. doi: 10.1126/science.298.5592.379. doi:10.1126/science.298.5592.379 [DOI] [PubMed] [Google Scholar]
- Narins P.M, Feng A.S, Yong H.S, Christensen-Dalsgaard J. Morphological, behavioral, and genetic divergence of sympatric morphotypes of the treefrog Polypedates leucomystax in Peninsular Malaysia. Herpetologica. 1998;54:129–142. [Google Scholar]
- Ron S.R, Santos J.C, Cannatella D.C. Phylogeny of the túngara frog genus Engystomops (Physalaemus pustulosus species group; Anura: Leptodactylidae) Mol. Phylogenet. Evol. 2006;39:392–403. doi: 10.1016/j.ympev.2005.11.022. doi:10.1016/j.ympev.2005.11.022 [DOI] [PubMed] [Google Scholar]
- Sodhi N.S, Koh L.P, Brook B.W, Ng P.K.L. Southeast Asian biodiversity: an impending disaster. Trends Ecol. Evol. 2004;19:654–660. doi: 10.1016/j.tree.2004.09.006. doi:10.1016/j.tree.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Stuart S.N, Chanson J.S, Cox N.A, Young B.E, Rodrigues A.S.L, Fischman D.L, Waller R.W. Status and trends of amphibian declines and extinctions worldwide. Science. 2004;306:1783–1786. doi: 10.1126/science.1103538. doi:10.1126/science.1103538 [DOI] [PubMed] [Google Scholar]
- Taylor E.H. The amphibian fauna of Thailand. Univ. Kansas Sci. Bull. 1962;43:265–599. [Google Scholar]
- Toda M, Matsui M, Nishida M, Ota H. Genetic divergence among Southeast and East Asian populations of Rana limnocharis (Amphibia: Anura), with special reference to sympatric cryptic species in Java. Zool. Sci. 1998;15:607–613. doi: 10.2108/0289-0003(1998)15[607:GDASAE]2.0.CO;2. doi:10.2108/zsj.15.607 [DOI] [PubMed] [Google Scholar]
- Wynn A, Heyer W.R. Do geographically widespread species of tropical amphibians exist? An estimate of genetic relatedness within the neotropical frog Leptodactylus fuscus (Schneider 1799) (Anura Leptodactylidae) Trop. Zool. 2001;14:255–285. [Google Scholar]
- Veith M, Kosuch J, Ohler A, Dubois A. Systematics of Fejervarya limnocharis (Gravenhorst, 1829) (Amphibia, Anura, Ranidae) and related species. 2. Morphological and molecular variation in frogs from the Greater Sunda Islands (Sumatra, Java, Borneo) with the definition of two species. Alytes. 2001;19:5–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Voucher information, sequencing protocols, GenBank accession numbers and methods of phylogenetic analyses