Abstract
During the Pleistocene pygmy elephantids, some only a quarter of their ancestors' size, were present on Mediterranean islands until about 10 000 years ago (y.a.). Using a new methodology for ancient DNA (aDNA) studies, the whole genomic multiple displacement amplification method, we were able to retrieve cytochrome b (cytb) DNA fragments from 4200 to 800 000 y.a. specimens from island and mainland samples, including pygmy and normal-sized forms. The short DNA sequence (43 bp) retrieved from the 800 000 y.a. sample is one of the oldest DNA fragment ever retrieved. Duplication of the experiments in two laboratories, the occurrence of three diagnostic sites and the results of the phylogenetic analyses strongly support its authenticity. Our results challenge the prevailing view that pygmy elephantids of the eastern Mediterranean originated exclusively from Elephas, suggesting independent histories of dwarfism and the presence of both pygmy mammoths and elephant-like taxa on these islands. Based on our molecular data, the origin of the Tilos and Cyprus elephantids from a lineage within the genus Elephas is confirmed, while the DNA sequence from the Cretan sample falls clearly within the mammoth clade. Thus, the name Mammuthus creticus rather than Elephas creticus, seems to be justified for this form. Our findings also suggest a need to re-evaluate the evolutionary history of the Sicilian/Maltese species, traditionally included in the genus Elephas.
Keywords: cytochrome b, elephantids, fossils, mammoths, mtDNA, taxonomy
1. Introduction
From the Late Pliocene onward, the Elephantidae was comprised of three principal lineages: Elephas, Mammuthus and Loxodonta. Throughout its evolutionary history, Loxodonta remained in Africa. Elephas dispersed twice out of Africa; once into Asia in the Mid-Pliocene, where it gave rise to Elephas maximus and once into Asia and Europe during the Late Pliocene as lineages which are now extinct (Elephas antiquus). Mammuthus migrated out of Africa in the Late Pliocene and subsequently spread rapidly throughout Europe, Asia and North America (Yang et al. 1996).
A noteworthy evolutionary trend in the Elephantidae is a reduction of body size in insular forms (Todd & Roth 1996). Populations of elephantids that colonized islands without large carnivores rapidly declined in body size, in some instances to a shoulder height of 1.5 m or less (Mol et al. 1996). This dwarfism is, in part, a consequence of natural selection for survival and reproduction in conditions of limited resources (Todd & Roth 1996). In the Mediterranean, their remains are known on the islands of both the western (Sardinia, Sicily, Malta and the Egad Islands) and eastern (Crete, Cyclades, Dodecanese, Cyprus) basins (Mol et al. 1996).
Despite extensive morphological research, the origin of the Mediterranean pygmy elephantids remains unclear. One hypothesis suggests that all the Mediterranean pygmy elephantids derived from E. antiquus, while the second maintains that in a few Mediterranean islands the pygmy elephantids are derivatives of Mammuthus (Mol et al. 1996). A previous molecular study (Poulakakis et al. 2002) suggested that the pygmies of Tilos (a Greek island) derived from Elephas rather than Mammuthus, supporting the first scenario. However, there is uncertainty about whether this is true for all pygmy elephantids from the Mediterranean since these elephantids are not contemporaneous. The elephantids of Tilos, Rhodes and Cyprus are Late Pleistocene, while those from Crete and Sardinia are early Middle Pleistocene.
In order to resolve the origin and evolutionary history of elephantids from some of the Mediterranean islands, we implemented molecular protocols based on whole genomic amplification (WGA, Dean et al. 2002; Mamone 2003) to generate fragments of the cytb mtDNA gene from DNA preserved in fossil bones and teeth. This is the first time this protocol has been applied to aDNA studies.
2. Material and methods
The ages of pygmy elephantids used in the study are in table 1. Standard aDNA handling conditions and dedicated equipment were used for sample collection, cleaning and DNA extractions (see Poulakakis et al. 2002). In an effort to increase the amount of aDNA retrieved we applied a WGA technique, multiple displacement amplification (MDA), using the GenomiPhi WGA Kit (Amersham). Following the first study, which implemented the MDA technique to increase the DNA quantity retrieved from human tissues (for clinical purposes), this method has been also applied in studies from efforts to increase DNA quantities from single DNA extractions in Insects (Gorrochotegui-Escalante & Black 2003) to molecular haplotyping (Paul & Apgar 2005).
Table 1.
Species (with species names as suggested therein), sample localities, source material for DNA extraction and accession numbers of specimens used in the study (only for haplotypes not in GenBank).
| species/subspecies | locality | material | age (y.a.) | acc no. |
|---|---|---|---|---|
| Elephas cypriotes | Cyprus (Akrotiri-Aetokremnos)a | vertebra (axis) | 10 000 | DQ329148 |
| Elephas maximus asurus | Iraq (Kish)b | molar | 4200 | DQ329149 |
| Elephas antiquus falconeri | Greece (Tilos, Charkadio cave)c | rib | 17 000 | DQ329147 |
| Mammuthus creticus | Greece (Crete, Cape Meleka)d | rib | ∼800 000 | —e |
Kourion Museum, Episkopi, Cyprus, FN136.
Department of Anthropology, The Field Museum, Chicago, FM 236234.
Natural History Museum of Crete, University of Crete (NHMC 20.2.2.1).
Natural History Museum of Crete, University of Crete (NHMC 20.1.2.30).
M. creticus (43 bp)-TACTATGGGTCCTACCTATACTCGGAAACCTGAAATACCGGCA.
A fragment of the cytochrome b (cyt b) was targeted using two pairs of specific primers (see electronic supplementary material). All procedures, except DNA sequencing, were carried out in facilities dedicated to ancient DNA studies and were done in duplicate (Yale University and University of Crete). No mammoth samples were ever located in either laboratory.
Analyses of the aDNA sequences from this study were carried out together with all published cyt b haplotypes from extinct and extant elephantid taxa available from GenBank (E. maximus, Loxodonta africana, Loxodonta cyclotis and Mammuthus primigenius). Population aggregation analysis (PPA; Davis & Nixon 1992) was used to identify diagnostic sites (i.e. fixed difference between predetermined lineages) for the two lineages (Elephas and Mammuthus) and to assign each aDNA sequence to a given taxon. Four outgroup taxa (electronic supplementary material) were used in the analysis.
Maximum Likelihood (ML) phylogenetic analysis was carried out on complete (40 taxa, 259 bp) and reduced (no missing data, 40 taxa, 43 bp) datasets (electronic supplementary material).
3. Results
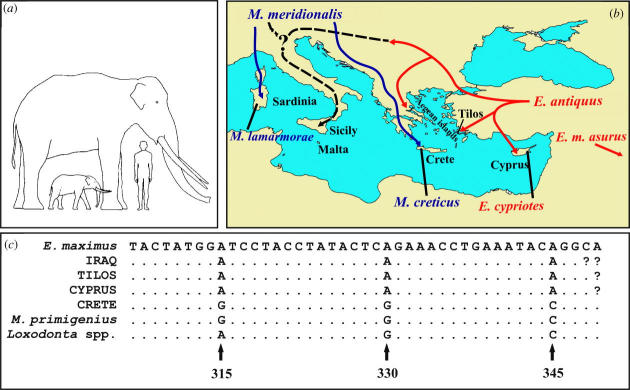
Multiple attempts with varying PCR conditions and nested PCR protocols did not result in any visible PCR products when we used the non-MDA treated DNA extractions for any of the samples. Using the MDA treated samples we were able to increase the overall amount of genomic material retrieved from the bone and teeth samples. This allowed the PCR amplification of fragments of the cytb gene ranging from 43 to 258 base pairs for three pygmy elephantid samples from Crete, Tilos and Cyprus and from an extinct normal-sized elephant from Iraq. PPA analyses assigned the Tilos, Cyprus and Iraq samples to Elephas and the Crete sample to Mammuthus underscoring the taxonomic importance of this small DNA cyt b fragment (figure 1c).
Figure 1.
(a) Pygmy and normal-sized elephantids next to a human for scale; (b) Proposed origin of Pleistocene pygmy elephantids on Mediterranean islands from Mammuthus (blue lines) and Elephas (red lines). Dashed lines indicate the possible origin of the elephantids from Sicily/Malta. (c) Alignment of cyt b Crete, Tilos, Cyprus and Iraq haplotypes with Elephas and Mammuthus consensus sequences. Dots and question marks identify DNA bases identical or missing (respectively) in comparison to the E. maximus sequence. Diagnostic sites (positions 315, 330 and 345 in E. maximus AB002412) are identified by arrows.
The ML analysis of the complete dataset resulted (figure 2) in a topology identical to that of the reduced dataset with ln L=−1116.4701 and very strong nodal support for the placement of the Cretan sample, formerly named Elephas creticus, within Mammuthus. The remaining Elephas samples were placed within the Elephas clade. Where appropriate, topological constraints were generated and compared with our optimal topology using the Shimodaira & Hasegawa (1999) SH-test. The assignment of the Cretan pygmy elephantid within the Mammuthus lineage was avouched by the results of the SH test (p<0.0001).
Figure 2.
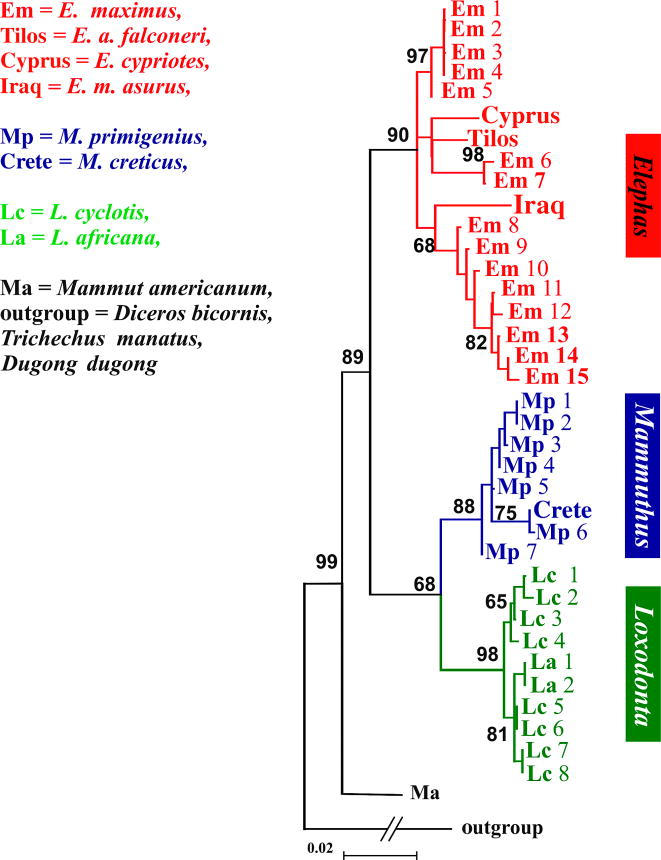
Cyt b phylogeny of all elephantids. Published haplotypes are indicated by a two-letter symbol and a number (see electronic supplementary material). The aDNA sequences produced for this study are indicated by the name of the locality of origin. Numbers on branches are bootstrap probabilities.
4. Discussion
The results were positive only after the MDA-treatment, making the Cretan sample one of the oldest aDNA fragments ever retrieved from bone samples. This technique might be an efficient way to overcome the problem of limited quantities of template DNA and to retrieve genetic information from samples for which standard aDNA extractions produce negative results. However, the drawbacks of the technique are: (i) its cost (approx. $5 per amplification) and (ii) the fact that is very sensitive, increasing the possibility of contamination by modern DNA. Nevertheless, its availability presents an opportunity to retrieve genetic information from a variety of degraded samples. Even though MDA appears to have many advantages for the recovery of DNA from sources of low-DNA quality and quantity, there have been as yet no systematic attempts to evaluate its use for the recovery of aDNA.
The value of MDA is evident in the case of the Cretan pygmy elephantid. It is known that the Pleistocene fauna of Crete included elephantids, hippos and deer. Both the pygmy elephantid and the pygmy hippopotamus were present on Crete during the Kritimys biozone (Middle Pleistocene). However, the pygmy elephantid lived during the Kritimys kiridus Subzone, while hippos (Hippopotamus creutzburgi) arrived on Crete during the Kritimys catreus Subzone. Three electron spin resonance dates for the Cretan hippo range from 846 000 to 475 550 y.a., while five amino acid racemization dates range from 738 000 to 378 000 y.a. (all ±20%; Reese et al. 1996). These dates and the consideration that biostratigraphically the pygmy elephantid was older than the pygmy hippopotamus suggest that the pygmy elephantid lived from the late Early Pleistocene to the early Middle Pleistocene (around 800 000 y.a.).
Although the DNA sequence from the Crete sample is only 43 bp, its taxonomic importance is underscored by the results of the PPA analysis, which assigned all cyt b sequences used in this study to either the Mammuthus or the Elephas clade (figure 1c), using three diagnostic sites in a 43 bp region. Two of them (sites 330 and 345 of E. maximus AB002412) are uniquely derived states (autapomorphies) for the Elephas clade, one (positions 315 of E. maximus AB002412) is a uniquely derived character state for the Mammuthus-Crete lineage (figure 1c).
This result is supported by the phylogenetic analysis, in which the cytb sequences of the two African elephants (L. africana and L. cyclotis), the extinct mastodon (Mammut americanum) and three more distantly related taxa are also included (figure 2). This analysis groups the aDNA sequences within two reciprocally monophyletic clades. One includes all Elephas haplotypes plus the ones from the Tilos, Cyprus and Iraq specimens, confirming previous data on Tilos samples (Poulakakis et al. 2002). The other comprises all Loxodonta and Mammuthus haplotypes, with the Crete DNA sequence firmly placed within the Mammuthus ones.
Although this analysis included missing data for the short Crete sequence, this should not impact our phylogenetic findings, since recent theoretical work showed that missing data present in a taxon does not limit the accuracy of its phylogenetic placement (Wiens 2003).
The inferred phylogenetic tree (figure 2) indicates that, depending on the geographic area, a different scenario is supported for the origin of the pygmy elephantids in the Mediterranean. Despite the small length of the amplified sequence, the relationship of the Cretan and the Tilos/Iraq lineage to the mammoths and the Asian elephants, respectively, cannot be disputed. The origin of the Tilos form from E. antiquus is confirmed, but the relationship of the Cretan form to the same ancestor is rejected.
Therefore, in light of the molecular data, the name Mammuthus creticus seems to be justified. Sardinia (Melis et al. 2001) is no longer the only island in the Mediterranean, where a pygmy mammoth existed.
When a new ancient DNA sequence is retrieved, it is often claimed that it will rewrite the textbooks of the organism it came from. This holds true for this study as well. This new data challenges the prevailing view that pygmy elephantids of the Mediterranean originated exclusively from Elephas, suggesting independent histories (figure 1b). The first (Middle Pleistocene) produced the Crete pygmy elephantids from the Mammuthus lineage. The second (Late Pleistocene) gave rise to the species on Cyprus and Tilos from Elephas. This scenario calls for a taxonomic revision of the Cretan form and a reconsideration of the origin of pygmy elephantids in the Mediterranean region. In fact, the phylogenetic history of the pygmy elephantids from Sicily/Malta has already been questioned (Lister & Bahn 1994). Morphology and stratigraphy suggest that the putative ancestor of these lineages (E. antiquus) is too recent for it to be the ancestor of the pygmy elephantids from Sicily/Malta. It is possible that the Sicilian/Maltese species also descended from Mammuthus rather than Elephas. If this is the case, then the western and eastern Mediterranean islands had different lineages, Mammuthus to the west and Elephas to the east of Crete (figure 1b).
Acknowledgments
This work was supported by a Niarchos Foundation grant to A.C. and E.Z. and we thank Armand Morgabn for the drawing in figure 1a.
Supplementary Material
References
- Davis J.I, Nixon K.C. Populations, genetic variation, and the delimitation of phylogenetic species. Syst. Biol. 1992;41:421–435. [Google Scholar]
- Dean F.B, et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. doi:10.1073/pnas.082089499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrochotegui-Escalante N, Black W.C. Amplifying whole insect genomes with multiple displacement amplification. Insect Mol. Biol. 2003;12:195–200. doi: 10.1046/j.1365-2583.2003.00401.x. doi:10.1046/j.1365-2583.2003.00401.x [DOI] [PubMed] [Google Scholar]
- Lister A, Bahn P. MacMillian; New York, NY: 1994. Mammoths. p. 168. [Google Scholar]
- Mamone J.T. A method for representatively amplifying genomic DNA. Genomic/Proteomic Technol. 2003;5:21–24. [Google Scholar]
- Melis R, Palombo M.R, Mussi M. Mammuthus lamarmorae (Major, 1883) remains in the pre-Tyrrhenian deposits of San Giovanni in Sinis (Western Sardinia; Italy) In: Cavarretta G, Gioia P, Mussi M, Palombo M.R, editors. The world of elephants. CNR; Rome, Italy: 2001. pp. 481–485. [Google Scholar]
- Mol D, DeVos J, van den Bergh G.D, Sondaar P.Y. The taxonomy and ancestry of the fossil elephants of Crete: faunal turnover and a comparison with Proboscidean faunas of Indonesian islands. In: Reese D.S, editor. Pleistocene and Holocene fauna of Crete and its first settlers. Prehistory Press; Madison, WI: 1996. pp. 81–98. [Google Scholar]
- Paul P, Apgar J. Single-molecule dilution and multiple displacement amplification for molecular haplotyping. Biotechniques. 2005;38:553–559. doi: 10.2144/05384ST01. [DOI] [PubMed] [Google Scholar]
- Poulakakis N, Theodorou G.E, Zouros E, Mylonas M. Molecular phylogeny of the extinct Pleistocene dwarf elephant Palaeoloxodon antiquus falconeri from Tilos island, Greece. J. Mol. Evol. 2002;55:364–374. doi: 10.1007/s00239-002-2337-x. doi:10.1007/s00239-002-2337-x [DOI] [PubMed] [Google Scholar]
- Reese D.S, Belluomini G, Ikeya M. Absolute dates for the pleistocene fauna of screte. In: Reese D.S, editor. Pleistocene and Holocene fauna of Crete and its first settlers. Madison; Madison, WI: 1996. pp. 47–51. [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- Todd N.E, Roth V.L. The proboscidea. Oxford University Press; Oxford, UK: 1996. Origin, species diversity and variability in the Elephantidae; pp. 193–202. [Google Scholar]
- Wiens J.J. Missing data, incomplete taxa, and phylogenetic accuracy. Syst. Biol. 2003;52:528–538. doi: 10.1080/10635150390218330. [DOI] [PubMed] [Google Scholar]
- Yang H, Golenberg E.M, Shoshani J. Phylogenetic resolution within the Elephantidae using fossil DNA sequence from the American mastodon (Mammut americanum) as an outgroup. Proc. Natl Acad. Sci. USA. 1996;93:1190–1194. doi: 10.1073/pnas.93.3.1190. doi:10.1073/pnas.93.3.1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.