Abstract
Population-based mapping approaches are attractive for tracing the genetic background to phenotypic traits in wild species, given that it is often difficult to gather extensive and well-defined pedigrees needed for quantitative trait locus analysis. However, the feasibility of association or hitch-hiking mapping is dependent on the degree of linkage disequilibrium (LD) in the population, on which there is yet limited information for wild species. Here we use single nucleotide polymorphism (SNP) markers from 23 genes in a recently established linkage map of the Z chromosome of the collared flycatcher, to study the extent of LD in a natural bird population. In most but not all cases we find SNPs within the same intron (less than 500 bp) to be in perfect LD. However, LD then decays to background level at a distance 1 cM or 400–500 kb. Although LD seems more extensive than in other species, if the observed pattern is representative for other regions of the genome and turns out to be a general feature of natural bird populations, dense marker maps might be needed for genome scans aimed at identifying association between marker and trait loci.
Keywords: collared flycatcher, recombination, Z chromosome
1. Introduction
Genomic research on humans and model organisms has increasingly come to consider using association (linkage disequilibrium, LD) or selective sweep (hitch-hiking) mapping as population-based alternatives to traditional, pedigree-based linkage mapping for the identification of genes or chromosomal regions underlying phenotypic traits (Kruglyak 1999). These approaches may very well find their application in future studies also of natural populations, in which there is considerable current interest in mapping and the subsequent study of the genetics of traits under selection in the wild (Feder & Mitchell-Olds 2003; Slate 2005). Given recent advances in large-scale identification and genotyping of single nucleotide polymorphisms (SNPs), association mapping in natural populations is about to become a realistic means for many organisms.
Critical to the feasibility of association mapping is the overall level of LD across the genome. When LD extends over large genomic regions, there is a higher chance of finding association between a gene affecting a particular phenotype and a linked marker at a given distance. On the other hand, long-range LD means that the physical distance between a gene and an associated marker can be significant, making gene identification less straightforward. Conversely, limited LD requires a much denser marker map for finding associations, however, when found, gene identification might be less tedious. Long-range LD can also be observed in cases where strong epistatic selection maintains associations among sites. To be able to make such inference, background levels of LD need to be known.
The potential for LD mapping in natural populations is thus dependent on how genetic diversity across the genome is structured in populations, about which there is so far almost no knowledge for the great majority of wild species. Benefiting from a recently established gene-based, high-density linkage map of the Z chromosome of the collared flycatcher (Ficedula albicollis; Backström et al. in press), we have now investigated the levels of LD in a natural bird population. The collared flycatcher is a highly philopatric, long-distance migrant with its main distribution in central and eastern Europe. It also occurs on the Baltic Sea islands Gotland and Öland with an estimated number of 5000 breeding pairs, where birds for this study were sampled. The species has been intensively studied in evolutionary research, including analyses of life history evolution, mate choice, sexual selection and speciation (e.g. Qvarnström et al. 2006). Our analysis now reveals that, on average, significant LD on the Z chromosome reaches up to 1 cM or 400–500 kb in this population.
2. Material and methods
Eighty-two female collared flycatchers from the Baltic Sea island Öland (n=70) and Gotland (n=12) sampled in 2001–2004 were analysed. These islands are located at a distance of 50 km from each other, with no significant evidence of population differentiation in our data (Fst=0.014; 95% confidence interval of −0.009–0.042). The sample included many mother–daughter pairs; since female birds inherit the single Z chromosome from their fathers, mother–daughter pairs present two unrelated Z chromosomes unless there is assortative mating on basis of Z chromosome genotypes or a strong role of Z-linked female preference genes for Z-linked male characteristics.
Intronic sequence data for male birds was obtained by PCR amplification using exonic primers designed from chicken genome sequence, followed by direct sequencing in both forward and reverse directions. Tentatively common SNPs identified by sequencing were subsequently genotyped in 82 female birds. Details of markers and protocols for sequencing and large-scale genotyping are given in Backström et al. (in press). For the purpose of this study, we used the observed gene order and recombination fractions as described in Backström et al. (in press) as genetic distance estimates between adjacent SNPs for addressing how LD decays with distance.
Estimates of LD measured as D′ and r2 were calculated in Haploview (Barrett et al. 2005); the use of female birds means that the Z chromosome phase is directly given. LD is preferably estimated using high-frequency polymorphisms (Reich et al. 2001) so we restricted the analysis to SNPs showing in the end a minor allele frequency (MAF) of at least 10%.
3. Results
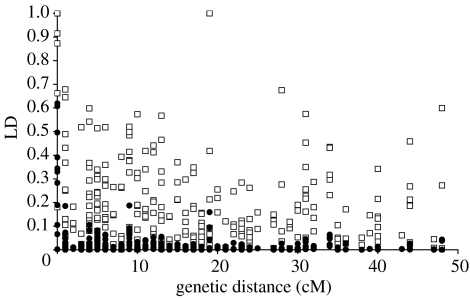
From re-sequencing of Z-linked introns in 8 male (16 chromosomes) collared flycatchers, we identified 34 high-frequency SNPs from 23 different genes spread over most of the Z chromosome. The mean distance between adjacent markers with detectable recombination was 2.9 cM (median 1.5 cM). After genotyping these SNPs in 82 female birds, significant levels of LD were found within most introns (mean, D′±s.d.=0.98±0.043; mean, r2=0.37±0.28), although there is evidence for recombination in two out of 12 pairwise comparisons from applying the four-gamete rule. LD then decays with increasing genetic distance (figure 1). For marker combinations with detectable recombination, the correlations between genetic distance and D′ (r2=0.0005) and r2 (r2=0.030) are very weak, indicating that a background level of LD is quickly reached.
Figure 1.
Plot of pair wise measures of D′ (open squares) and r2 (solid dots) versus genetic distance along the collared flycatcher Z chromosome.
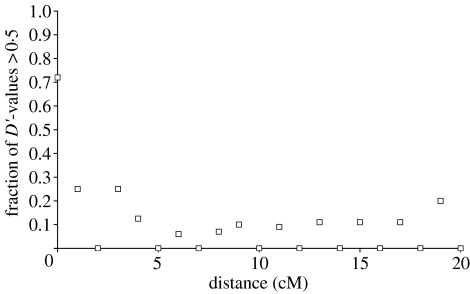
The ‘half-length’ of LD, defined either as the distance at which mean D′ drops below 0.5 or the distance at which the fraction of D′ values being less than 0.5 drops below 0.5, is a measure of how far significant LD reaches (Reich et al. 2001). A plot of the fraction of D′ values less than 0.5 in relation to genetic distance indicates that this half-length is less than 1 cM in our collared flycatcher sample (figure 2). If we assume an average recombination rate of 2 cM Mb−1 on the Z chromosome (Backström et al. in press), this indicates that significant LD extends no further than 500 kb.
Figure 2.
Fraction of D′ values exceeding 0.5 for the interval 0–20 cM.
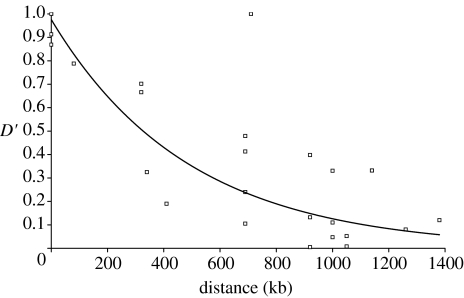
To address this in some further detail, we plot D′ in relation to the physical distance of adjacent markers in the interval 0–1000 kb, as given by the distance in orthologous sequence of the chicken genome (figure 3). Clearly, this can only be seen a crude approximation given that we do not know to what extent intergenic distances are conserved between collared flycatcher and chicken in these orthologous regions (intron size is indeed correlated: r2=0.52, p<0.01). However, the linkage map indicates that these regions are at least not involved in any large-scale rearrangements. Assuming that chicken and collared flycatcher physical distances are correlated, or at least not biased in any particular direction, this indicates that, on average, D′ drops below 0.5 at roughly 400 kb.
Figure 3.
D′ values for intergenic pairs of SNP markers located within a physical distance of 0–1000 kb in the chicken genome.
4. Discussion
Levels of LD in the genomes of natural populations of birds are largely unknown, with the notable exception of the detailed analysis by Edwards & Dillon (2004) concerning a 40 kb region of the MHC class II locus in red-winged blackbirds (Agelaius phoeniceus). They found high LD only across a few hundred base pairs but the generality of this observation is unclear given the characteristics of MHC loci, which are under the influence of balancing selection (Meyer & Thomson 2001), epistatic interactions and recombination hot spots (e.g. Kauppi et al. 2003). Moreover, the observation of significant LD extending over several centimorgans in commercial populations of domestic chicken may have limited relevance for the situation in natural bird populations (Heifetz et al. 2005). Other studies have indicated that LD decays rapidly, typically within 1 kb, in natural populations of e.g. maize (Remington et al. 2001), Plasmodium falciparium (Conway et al. 1999), and even of wild barley despite a high rate of self-fertilization in this species (Morrell et al. 2005). In the selfing plant Arabidopsis thaliana, LD extends up to 50–100 kb (Nordborg et al. 2005), and similarly long ranges are seen among humans (Reich et al. 2001). In Drosophila melanogaster LD generally reaches over very short distances (Andolfatto & Przeworski 2000), although there is significant variation in extent of LD among populations (Haddrill et al. 2005). Compared to these observations, levels of LD on the collared flycatcher Z chromosome seem high.
In theory, the expected degree of LD between alleles is given by the age of mutations and the rate of recombination between loci (Stumpf & McVean 2003). The key parameter is the population recombination rate (ρ), i.e. the product of the rate of recombination per generation (r) and the effective population size (Ne; ρ=4Ner). Moreover, since Ne will be dependent on e.g. population history, population subdivision and selection, a number of evolutionary forces are involved. Based on studies of the domestic chicken, it has been suggested that birds have high-recombination rates, a situation that would act against the build-up of extensive LD. The chicken genome project (ICGSC 2004) estimated the rate to be 5.4 cM Mb−1 for the Z chromosome. However, high-recombination rates may not be true for birds in general. The length of the map of the collared flycatcher Z chromosome is only about 50% of the length of the syntenic region of chicken Z (Backström et al. in press). Moreover, preliminary observations from a partial microsatellite-based linkage map of another passerine bird, the great reed warbler (Acrocephalus arundinaceus), suggest a rate only 17–32% of that in chicken (Hansson et al. 2005). Still, the rate of recombination in birds seems sufficiently high to, in principal, hinder the build-up of extensive LD.
The observed high levels of LD in collared flycatchers suggest low Ne, for instance owing to a population history involving bottlenecks associated with the re-colonization of isolated Baltic Sea island. Historical records indicate that collared flycatchers were not present on Gotland until in the mid 19th century (Alatalo et al. 1990), although this is based on reports from occasional expeditions only. However, if correct, re-colonization may have implied founder effects that still leave an imprint on the current patterns of LD in the population. This possibility should be further investigated by analysis of autosomal loci, and by comparisons with continental populations. However, the failure to detect significant departure from a neutral model in Tajima's D-tests (data not shown) is not easily compatible with a model of a bottleneck followed by recent population expansion.
Borge et al. (2005) found levels of genetic diversity at nine Z-linked loci in a sample of nine south European collard flycatchers to be lower than expected based on the observed polymorphism level at autosomal loci and assuming a neutral model. They argued that the Z chromosome might be low in genetic diversity because recurrent selective sweeps, caused e.g. by sexual selection, would reduce levels of polymorphism at linked loci. Indeed, the Z chromosome is likely to be particularly sensitive to sexual selection in systems of female heterogamety (e.g. Reeve & Pfennig 2003). Our finding of LD generally not extending over more than 500 kb on the collared flycatcher Z chromosome would suggest that, if selective sweeps have had a strong role in shaping the overall levels of Z chromosome diversity, strong positive selection must have occurred quite frequently across a large number of loci on the Z chromosome. This might be seen as an unlikely scenario.
Firmly establishing the magnitude of recombination rate variation among birds, and within avian genomes, will be an important aspect of future studies aimed at addressing LD in natural populations. However, if the pattern observed in this study holds true for most of the genome, it is indicated that association mapping will become a challenging task given the high density of markers needed to scan the genome. If mean LD extends ca 500 kb, roughly 1000–2000 evenly spaced markers would be required for covering most genomic regions in an association scan. While indeed a large number by present standard for most wild species, it should not be seen as unrealistic given current technological developments.
Acknowledgments
Financial support was obtained from the Swedish Research Council. The useful comments of two anonymous referees are acknowledged.
References
- Alatalo R.V, Eriksson D, Gustafsson L, Lundberg A. Hybridisation between pied and collared flycatchers—sexual selection and speciation theory. J. Evol. Biol. 1990;3:375–389. doi:10.1046/j.1420-9101.1990.3050375.x [Google Scholar]
- Andolfatto P, Przeworski M. A genome-wide departure from the standard neutral model in natural populations of Drosophila. Genetics. 2000;156:257–268. doi: 10.1093/genetics/156.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström, N., Brandström, N., Gustafsson, L., Qvarnström, A., Cheng, H. & Ellegren, H. In press. Genetic mapping in a natural population of collared flycatchers (Ficedula albicollis): conserved synteny but gene order rearrangements on the avian Z chromosome. Genetics [DOI] [PMC free article] [PubMed]
- Barrett J.C, Fry B, Maller J, Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. doi:10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Borge T, Webster M.T, Andersson G, Saetre G.P. Contrasting patterns of polymorphism and divergence on the Z chromosome and autosomes in two Ficedula flycatcher species. Genetics. 2005;171:1861–1873. doi: 10.1534/genetics.105.045120. doi:10.1534/genetics.105.045120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway D.J, Roper C, Oduola A.M, Arnot D.E, Kremsner P.G, Grobusch M.P, Curtis C.F, Greenwood B.M. High recombination rate in natural populations of Plasmodium falciparum. Proc. Natl Acad. Sci. USA. 1999;96:4506–4511. doi: 10.1073/pnas.96.8.4506. doi:10.1073/pnas.96.8.4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S.V, Dillon M. Hitchhiking and recombination in birds: evidence from MHC-linked and unlinked loci in red-winged blackbirds (Agelaius phoeniceus) Genet. Res. 2004;84:175–192. doi: 10.1017/s0016672304007189. doi:10.1017/S0016672304007189 [DOI] [PubMed] [Google Scholar]
- Feder M.E, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nat. Rev. Genet. 2003;4:651–657. doi: 10.1038/nrg1128. doi:10.1038/nrg1128 [DOI] [PubMed] [Google Scholar]
- Haddrill P.R, Thornton K.R, Charlesworth B, Andolfatto P. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 2005;15:790–799. doi: 10.1101/gr.3541005. doi:10.1101/gr.3541005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson B, Akesson M, Slate J, Pemberton J.M. Linkage mapping reveals sex-dimorphic map distances in a passerine bird. Proc. R. Soc. B. 2005;272:2289–2298. doi: 10.1098/rspb.2005.3228. doi:10.1098/rspb.2005.3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz E.M, Fulton J.E, O'Sullivan N, Zhao H, Dekkers J.C, Soller M. Extent and consistency across generations of linkage disequilibrium in commercial layer chicken breeding populations. Genetics. 2005;171:1173–1181. doi: 10.1534/genetics.105.040782. doi:10.1534/genetics.105.040782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. doi:10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Kauppi L, Sajantila A, Jeffreys A.J. Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum. Mol. Genet. 2003;12:33–40. doi: 10.1093/hmg/ddg008. doi:10.1093/hmg/ddg008 [DOI] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 1999;22:139–144. doi: 10.1038/9642. doi:10.1038/9642 [DOI] [PubMed] [Google Scholar]
- Meyer D, Thomson G. How selection shapes variation of the human major histocompatibility complex: a review. Ann. Hum. Genet. 2001;65:1–26. doi: 10.1046/j.1469-1809.2001.6510001.x. doi:10.1046/j.1469-1809.2001.6510001.x [DOI] [PubMed] [Google Scholar]
- Morrell P.L, Toleno D.M, Lundy K.E, Clegg M.T. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc. Natl Acad. Sci. USA. 2005;102:2442–2447. doi: 10.1073/pnas.0409804102. doi:10.1073/pnas.0409804102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. doi:10.1371/journal.pbio.0030196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnström A, Brommer J.E, Gustafsson L. Testing the genetics underlying the co-evolution of mate choice and ornament in the wild. Nature. 2006;441:84–86. doi: 10.1038/nature04564. [DOI] [PubMed] [Google Scholar]
- Reeve H.K, Pfennig D.W.P. Genetic biases for showy males: are some genetic systems especially conductive to sexual selection? Proc. Natl Acad. Sci. USA. 2003;100:1089–1094. doi: 10.1073/pnas.0337427100. doi:10.1073/pnas.0337427100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D.E, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. doi:10.1038/35075590 [DOI] [PubMed] [Google Scholar]
- Remington D.L, Thornsberry J.M, Matsuoka Y, Wilson L.M, Whitt S.R, Doebley J, Kresovich S, Goodman M.M, Buckler E.S., IV Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc. Natl Acad. Sci. USA. 2001;98:11 479–11 484. doi: 10.1073/pnas.201394398. doi:10.1073/pnas.201394398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 2005;14:363–379. doi: 10.1111/j.1365-294X.2004.02378.x. doi:10.1111/j.1365-294X.2004.02378.x [DOI] [PubMed] [Google Scholar]
- Stumpf M.P, McVean G.A. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 2003;4:959–968. doi: 10.1038/nrg1227. doi:10.1038/nrg1227 [DOI] [PubMed] [Google Scholar]