Abstract
The proper timing of flowering is critical for successful reproduction. The perception of the seasonal cues of day-length changes and exposure to cold influences flowering time in many plant species through the photoperiod and vernalization pathways, respectively. Here we show that a plant homeodomain (PHD) finger-containing protein, VIN3-LIKE 1 (VIL1), participates in both the photoperiod and vernalization pathways in Arabidopsis thaliana by regulating expression of the related floral repressors FLOWERING LOCUS C (FLC) and FLOWERING LOCUS M (FLM). In the vernalization pathway, VIL1, along with VERNALIZATION INSENSITIVE 3 (VIN3), is necessary for the modifications to FLC and FLM chromatin that are associated with an epigenetically silenced state and with acquisition of competence to flower. In addition, VIL1 regulates FLM independently of VIN3 in a photoperiod-dependent manner.
Keywords: Epigenetics, flowering, histone modification, photoperiod, vernalization
The transition to flowering is a critical developmental event in the plant life cycle. In many plants, the perception of seasonal changes affects the timing of this transition. Two environmental cues that many plant species monitor for flowering time control are the prolonged cold of winter and changing day length; these cues promote flowering through the vernalization and photoperiod pathways, respectively. Most accessions of Arabidopsis thaliana exhibit a facultative response to these signals: In the absence of either environmental cue, flowering is delayed but nevertheless eventually occurs.
In Arabidopsis, vernalization results in the mitotically stable repression of the potent floral repressor, FLOWERING LOCUS C (FLC) (Michaels and Amasino 1999; Sheldon et al. 1999). Vernalization-mediated repression of FLC is associated with histone modifications such as methylation of histone H3 Lys 9 (H3K9) and Lys 27 (H3K27) as well as H3 deacetylation (Bastow et al. 2004; Sung and Amasino 2004). A plant homeodomain (PHD) finger-containing protein, VERNALIZATION INSENSITIVE 3 (VIN3), is required for FLC repression by vernalization and the associated modifications to FLC chromatin (Sung and Amasino 2004). Here we show that a related PHD finger-containing protein, VIN3-LIKE 1 (VIL1), identified in a screen for proteins that interact with VIN3, cooperates with VIN3 in the vernalization-mediated repression of FLC. Furthermore, independently of VIN3 activity, VIL1 mediates the photoperiod-specific repression of another member of the FLC clade. Thus, VIL1 is involved in the regulation of flowering by two environmental-sensing pathways.
Results and Discussion
Because VIN3 is essential for the vernalization response (Sung and Amasino 2004) and PHD finger-containing proteins are often members of multisubunit chromatin remodeling complexes (Bienz 2006), we searched for potential components of a VIN3 complex using the yeast two-hybrid system. This screen revealed two independent VIN3-interacting clones encoding C-terminal regions of At3g24440. Interestingly, At3g24440 encodes a protein similar to VIN3, and thus we designated At3g24440 as VIL1. VIN3 and VIL1 are members of an Arabidopsis clade that encodes proteins sharing sequence and domain structure similarities (Fig. 1A; Supplementary Fig. S1). All members of the VIN3 clade have a conserved C-terminal domain, which to date is unique to the plant kingdom.
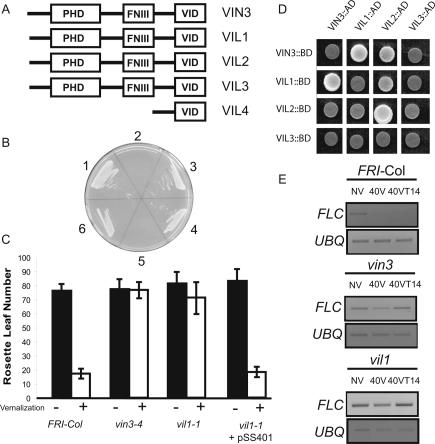
Figure 1.
(A) Domain structures of VIN3-like proteins. (Amino acid alignments are provided in Supplementary Fig. 1.). VIN3 is At5g57380; VIL1 is At3g24440; VIL2 is At4g30200; VIL3 is At2g18880; and VIL4 is At2g18870. (B) The VID region is required for the interaction between VIN3 and VIL1. The constructs either have the VID region alone as described in Supplementary Figure S1 or the remainder of the gene without the VID region. Yeast two-hybrid assays: (1) full-length VIN3 as bait and full-length VIL1 as a prey. (2) VIN3 without VID as bait and full-length VIL1 as a prey. (3) VID of VIN3 as bait and full-length VIL1 as prey. (4) Full-length VIL1 as bait and full-length VIN3 as prey. (5) Full-length VIL1 as bait and VIN3 without VID as prey. (6) Full-length VIL1 as bait and VID of VIN3 as prey. Yeast two-hybrid assay conditions are described in Materials and Methods (Supplemental Material). (C) Yeast two-hybrid assays among the VID regions. (D) Vernalization response in vil1 and vin3. (Filled bars) Leaf number at flowering of nonvernalized plants; (open bars) values for plants cold-treated for 40 d as described in Materials and Methods (Supplemental Material). (E) FLC mRNA levels in vin3 and vil1. (NV) Samples from nonvernalized plants grown at 22°C; (40V) samples prepared directly from plants grown for 40 d at 4°C; (40VT14) plants grown for 40 d at 4°C followed by growth at 22°C for 14 d.
That the screen identified clones containing only the conserved C-terminal domain of VIL1 indicated that this region is sufficient to mediate the interaction with VIN3. To delineate the regions of VIN3 that interact with the C-terminal domain of VIL1, we performed two-hybrid assays using a series of VIN3 deletions. This analysis revealed that the conserved C-terminal domain of VIN3 is necessary and sufficient for the two-hybrid interaction (Fig. 1B). We refer to this C-terminal domain as the VIN3-Interacting Domain (VID). Furthermore, pairwise examination of the interactions among the VID regions of VIN3 and other family members indicated that, in addition to the VIN3:VIL1 interaction, VIL2 interacts strongly with itself and weakly with VIN3 and VIL1 (Fig. 1C).
To evaluate whether VIL1 plays a role in vernalization, we isolated two T-DNA insertion mutants (SALK_136506, vil1-1; SALK_140132, vil1-2) and introduced these vil1 alleles into the vernalization-requiring genetic background FRI-Col (the Columbia accession into which FRIGIDA has been introgressed as described in Lee and Amasino [1995). VIL1 is indeed required for vernalization: vil1 mutants exhibit an impaired vernalization response similar to that of vin3 (Fig. 1D). Furthermore, lesions in vin3 and vil1 also cause the similar molecular phenotypes of incomplete repression of FLC during cold exposure and a lack of FLC repression after cold exposure (Fig. 1E).
A molecular phenotype of vernalization is an enrichment of H3K9 and H3K27 dimethylation at FLC chromatin (Bastow et al. 2004; Sung and Amasino 2004). However, for the following reasons, we evaluated whether H3K9 and H3K27 trimethylation of FLC chromatin might also be associated with the vernalized, repressed state. First, LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), the plant homolog of HETEROCHROMATIN PROTEIN 1 (HP1), is associated with FLC chromatin after vernalization (Sung et al. 2006). On a genome-wide level, LHP1 localizes predominantly to regions of euchromatin (Libault et al. 2005; Nakahigashi et al. 2005; Sung et al. 2006), and a recent study indicates that in plants, trimethylated H3K27 also localizes to regions of euchromatin (Mathieu et al. 2005). Second, trimethylated H3K9 is more specifically associated with repressed genes that are bound by HP1 than is dimethylated H3K9 (Jacobs and Khorasanizadeh 2002). Vernalization results in an enrichment of both H3K9 and H3K27 trimethylation at FLC chromatin in a pattern similar to the changes in dimethylation (Fig. 2B–E). Notably, regions at which trimethylation of H3K9 and H3K27 is enriched in FLC chromatin (Fig. 2D,E; Supplementary Fig. S2) are correlated with regions at which LHP1 is also enriched by vernalization (Sung et al. 2006). Thus, although at a genome-wide level trimethylation of H3K9 and H3K27 exists at low levels in Arabidopsis (Johnson et al. 2004), these modifications may play a key role in the repression of developmentally regulated genes in Arabidopsis.
Figure 2.
Chromatin immunoprecipitation (ChIP) analyses of vernalization-mediated histone modifications in wild type and vil1. Samples are from plants grown as follows: nonvernalized (NV); vernalized for 40 d (40V); vernalized for 40 d and subsequently grown for 14 d at 22°C (40VT14). Relative fold changes are indicated at the bottom of each ChIP assay. All assays were repeated with at least two independent chromatin preparations and three independent immunoprecipitations as described in Materials and Methods. Representative ChIP results are shown using antibodies recognizing acetylated Histone H3 (A), dimethylated Histone H3 Lys 9 (B), dimethylated Histone H3 Lys 27 (C), trimethylated Histone H3 Lys 9 (D), and trimethylated Histone H3 Lys 27 (E). The regions of FLC and FLM chromatin examined are described in Supplementary Figure S2.
In a vil1 mutant, the histone methylations characteristic of the vernalized state of FLC chromatin do not occur (Fig. 2). Thus, that the physical interaction between VIN3 and VIL1 observed in a two-hybrid assay is critical for vernalization is consistent with the fact that both vin3 and vil1 mutants exhibit similar phenotypes including an impaired flowering response to a vernalizing cold treatment as well as an inability to initiate the modifications to FLC chromatin associated with vernalization.
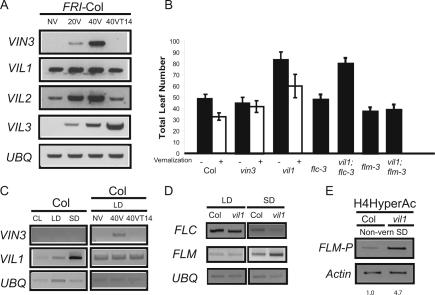
VIN3 is only expressed in response to long periods of cold exposure (Fig. 3A; Sung and Amasino 2004); however, cold exposure has little effect on VIL1 mRNA levels. In short days (SD; 8 h light/16 h dark), VIL1 is highly expressed before, during, and after cold exposure with perhaps only a slight increase in mRNA levels during cold exposure (SD are standard conditions to evaluate the effect of cold exposure on mRNA levels because vernalization typically occurs during the SD of winter) (Fig. 3A). The temperature-independent expression of VIL1 indicates it may have other roles in flowering in addition to its role in vernalization. Therefore, we evaluated the flowering behavior of vil1 mutants in a variety of conditions. A vil1 lesion does not affect the flowering behavior of nonvernalized FRI-Col grown in inductive long days (LD; 16 h light/8 h dark) (Fig. 1D), nor does it affect flowering in LD in the wild-type Col background (i.e., Col that does not have an active FRI allele) (Johanson et al. 2000). However, vil1 mutants exhibit a substantial delay of flowering in SD (Fig. 3B; data shown for Col lines without an active FRI allele). Thus, in addition to being required for vernalization, VIL1 promotes flowering in SD. This is in contrast to the vernalization-specific role of VIN3; vin3 lesions do not alter the timing of flowering in either LD or SD in nonvernalized plants (Figs. 1D, 3B; Sung and Amasino 2004).
Figure 3.
(A) Effect of cold exposure on mRNA levels of VIN3 family members. (B) SD flowering behavior. (Filled bars) Leaf number at flowering of nonvernalized plants; (open bars) leaf number of plants vernalized for 40 d as described in Materials and Methods (Supplemental Material). (C, left) mRNA levels of VIN3 and VIL1 in different photoperiod conditions. (CL) Continuous light; (LD) long days, 16 h light/8 h dark; (SD) short days, 8 h light/16 h dark. (Right) mRNA levels of VIN3 and VIL1 during vernalization under LD. Samples were nonvernalized (NV), vernalized for 40 d (40V), and vernalized for 40 d and subsequently grown for 14 d at 22°C (40VT14). (D) mRNA levels of FLC and FLM in wild-type Columbia (Col) and vil1 mutants grown in LD and SD. (The stronger FLC signal in LD is a result of intentional dilution of the cDNA templates from SD samples relative to LD samples to facilitate evaluation of the increase in FLM in SD-grown vil1 mutants.) (E) ChIP assay using antibodies recognizing hyperacetylated Histone H4. Chromatin was prepared from wild-type (Col) and vil1 mutants grown in SD without vernalization treatment. Three independent immunoprecipitation were performed, and a representative result is shown. Relative fold changes are indicated at the bottom of each ChIP assay.
Because VIL1 promotes flowering in SD, we examined whether VIL1 expression was photoperiod regulated. VIL1 is expressed at higher levels in SD than in LD (Fig. 3C), consistent with its role in promoting flowering in SD. Because VIL1 expression is lower in LD, we also evaluated whether cold exposure might induce VIL1 in LD, but VIL1 is not induced during vernalization in these conditions (Fig. 3C). Thus, unlike VIN3, which is regulated by cold exposure regardless of photoperiod, the expression of VIL1 is photoperiod regulated, but not cold regulated (Fig. 3A,C).
Despite the different environmental signals that affect VIN3 and VIL1 expression, both VIN3 and VIL1 are required for the vernalization-mediated silencing of FLC (Fig. 2B). We therefore investigated whether the delayed flowering of a nonvernalized vil1 mutant in SD is due to altered FLC expression. The levels of FLC expression in wild type and vil1 are the same regardless of photoperiod (Fig. 3D; as noted in the legend, the SD samples were diluted relative to LD samples to emphasize the FLM mRNA difference). Furthermore, loss of FLC does not suppress the late-flowering phenotype of vil1 mutants in SD (Fig. 3B).
FLC belongs to a clade of genes in Arabidopsis (other members: FLOWERING LOCUS M [FLM]/MADS AFFECTING FLOWERING 1 [MAF1], MAF2, MAF3, MAF4, and MAF5) in which certain members, particularly FLM, repress flowering (Ratcliffe et al. 2001; Scortecci et al. 2001). Altered FLM expression appears to be responsible for the vil1 SD phenotype. Among the members of the FLC clade, only FLM is up-regulated in vil1 mutants in SD compared with the wild type (Fig. 3D; data shown for FLM), but in LD, conditions in which loss of vil1 does not affect the flowering of nonvernalized plants, there is no effect of the vil1 lesion on FLM levels (Fig. 3D). Moreover, loss of FLM activity eliminates the late-flowering phenotype of the vil1 lesion in SD (Fig. 3B). Consistent with the enhanced FLM expression in a vil1 mutant, FLM chromatin is more highly acetylated in vil1 (Fig. 3E). Thus, VIL1, in addition to its role in vernalization, functions in the photoperiodic regulation of flowering. Specifically, the VIL1-mediated repression of FLM in SD serves to attenuate the photoperiod response. In the absence of VIL1 activity, FLM mRNA levels are much higher in SD relative to LD, but in wild type, VIL1 attenuates the photoperiod effect on FLM expression. The up-regulation of VIL1 in SD (Fig. 3C) may be necessary for full attenuation of FLM.
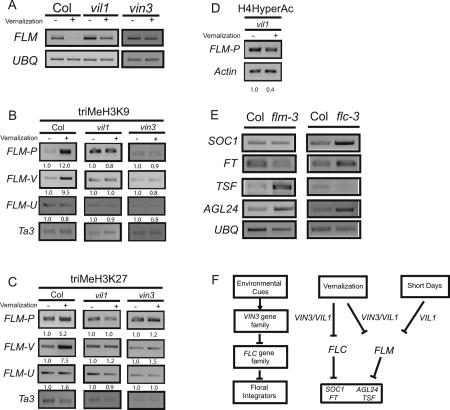
The additional delay of flowering in a vil1 mutant in SD is reversed by vernalization (Fig. 3B). At a molecular level, vernalization reduces the elevated expression of FLM in vil1 to the level of nonvernalized wild type, but does not reduce FLM expression as completely as that in vernalized wild type (Fig. 4A; note comment on vernalization-mediated FLM repression in legend). Thus, as shown above for FLC, VIL1 is required for the complete vernalization-mediated repression of FLM. To investigate the molecular basis of FLM repression by vernalization, we examined covalent modifications of FLM chromatin during and after cold exposure. Similar to that observed for FLC chromatin (Fig. 2B,C), in wild type, vernalization results in a stable increase in H3K9 and H3K27 trimethylation of FLM chromatin, but these modifications do not occur in vil1 or vin3 mutants (Fig. 4B,C). Thus, both VIN3 and VIL1 are required for the vernalization-mediated increase in tri- H3K9 and H3K27 methylation of FLM chromatin. As noted above, there is an additional increment of FLM expression that occurs in SD in a vil1 mutant that can be repressed by vernalization without VIL1 activity (Fig. 4A) and without an apparent increase in the H3K9 and H3K27 trimethylation of FLM chromatin (Fig. 4B,C). In a vil1 mutant background, however, there is a vernalization-mediated decrease in levels of H4 acetylation of FLM chromatin (Fig. 4D). Thus, although VIL1 contributes to FLM repression in SD and to the strong silencing of FLM by vernalization, there is a VIL1-independent mechanism that can reduce the ectopic FLM expression of a vil1 mutant during vernalization.
Figure 4.
(A) Effect of vernalization on mRNA levels of FLC and FLM in wild-type (Col) and vil1 mutants grown in SD. Samples in all panels unless noted are nonvernalized (−) or cold-treated for 40 d and subsequently grown for 10 d at 22°C under SD (+). (Note: In Scortecci et al. [2001], we reported that FLM mRNA levels were not reduced by vernalization in a FRI-containing line. However, we now reproducibly detect a vernalization-mediated decrease of FLM with or without FRI.) (B) ChIP assays using antibodies recognizing trimethylated Histone H3 Lys 9 at FLM chromatin in wild type (Col), vin3, and vil1. (C) ChIP assay using antibodies recognizing trimethylated Histone H3 Lys 27 at FLM chromatin in wild type (Col), vin3, and vil1. (D) ChIP assay using antibodies recognizing hyperacetylated Histone H4 at FLM chromatin in vil1 mutants. (E) mRNA expression patterns of floral integrator genes in wild-type (Col), flm, and flc mutants grown in SD. cDNA templates prepared from mutant seedlings were intentionally diluted relative to wild-type samples to emphasize the increased mRNA levels in mutants. (F) Relationship of VIN3 family genes to the regulatory network controlling flowering time in response to environmental cues.
Although both FLC and FLM function as floral repressors, FLC appears to have a greater role than FLM in repressing SOC1 and FT (Fig. 4E), consistent with previous reports that both SOC1 and FT targets are bound directly by FLC (Helliwell et al. 2006; Searle et al. 2006). Interestingly, the mRNA levels of TWIN SISTER OF FT (TSF), a relative of FT that also promotes flowering (Michaels et al. 2005; Yamaguchi et al. 2005), are more affected by loss of FLM than by loss of FLC (Fig. 4E). Thus, there appears to be specialization in the roles of FLC and FLM in repressing specific floral activators. This specialization, along with the role of VIL1 but not VIN3 in the photoperiodic control of flowering, illustrates the intricate regulatory network between the VIN3 and the FLC gene families and downstream “integrators” of flowering pathways (Fig. 4F).
In conclusion, VIN3 and VIL1 are required for the vernalization-mediated changes in the chromatin structure of FLC and FLC relatives like FLM that lead to the establishment of competence to flower characteristic of the vernalized state. VIL1 has the additional role in the photoperiodic regulation of flowering time of attenuating the level of FLM expression in noninductive photoperiods. Thus, in Arabidopsis, two members of the VIN3 family have evolved to transduce perception of the environmental cues of a vernalizing cold treatment and SD photoperiods into regulation of target genes. The VIN3 gene family is found in other plant species, and it will be interesting to evaluate whether VIN3 family members are involved in the flowering response to environmental signals in other species.
Materials and methods
Plants materials
T-DNA insertion mutants for At3g24440 were obtained from Arabidopsis Biological Resource Center (ABRC) (Alonso et al. 2003), and a homozygous line for SALK_136506 was designated as vil1-1 and SALK_140132 as vil1-2. vil1-1 and vil1-2 in Columbia were introgressed into FRI-Col (Lee and Amasino 1995). A T-DNA insertion mutant for FLM (SALK_141971) was also obtained from ABRC (Alonso et al. 2003), and a homozygous line was designated as flm-3. flc-3 was previously described (Michaels and Amasino 1999).
Plant transformation
The coding region of VIL1 was first subcloned into the pENTR directional TOPO vector (Invitrogen) followed by LR reaction (Invitrogen) into the destination vector pEarleyGate100 (Earley et al. 2006) using LR clonase enzyme (Invitrogen). The resulting construct was designated pSS401 and transformed into Agrobacterium and used for plant transformation. Plant transformations were done as described (Clough and Bent 1998).
Yeast two-hybrid assays
cDNAs were amplified using RT–PCR and subcloned into the pENTR directional TOPO vector (Invitrogen) followed by LR reaction (Invitrogen) into destination vector pDEST32 and pDEST22 using LR clonase enzyme (Invitrogen). Yeast transformation and analyses were performed using the ProQuest Two-Hybrid System with Gateway Technology (Invitrogen). Interactions were verified by growing yeasts on plates with SC−Trp−Leu−His + 3AT (100 mM).
Vernalization treatment
Seeds were germinated on agar plates for 5 d at 22°C and vernalized for 40 d at 4°C under 8 h of light and 16 h of dark unless indicated otherwise. Post-vernalization samples continued to grow on agar plates under 8 h of light and 16 h of dark at 22°C. All plants samples were prepared at 10 a.m. (2 h after lights on).
Chromatin immunoprecipitation (ChIP) analysis
Chromatin samples were prepared as described (Gendrel et al. 2005). Immunoprecipitations were performed using a ChIP assay kit (Upstate Biotechnology; no. 17-295) following the manufacturer’s suggested protocol modified as previously described (Johnson et al. 2002). Antibodies against trimethyl H3K9, trimethyl H3K27, dimethyl H3K9, dimethyl H3K27, and hyperacetylated H4 were obtained from Upstate Biotechnology. Primers used to detect Ta3, actin, and FLC-P, FLC-V, and FLC-U regions were previously described (Sung et al. 2006). The primers used to detect FLM-P were 5′-TTAAATTAGGGCATAACCCTTATCGGAGA-3′ and 5′-GAGAATCGAAAGTTGTCGAGCTTTGTCGA-3′; to detect FLM-V, 5′-CCACTAGACTACAACCAAATGTTCAATGA-3′ and 5′-TGGAGTCAACATCAAGGAAATGAGAAGGA-3′; and to detect FLM-U 5′-GCGTGAAGAACCCTAGACCTTCTTATTTC-3′ and 5′-AA CATGAAACAGAGTTCAAAAGCTGGCTT-3′. Fold changes were calculated compared with controls using ImageQuant. All ChIP assays were performed at least three times from at least two chromatin samples prepared from biological replicates. It should be noted that unlike ChIP using antibodies against dimethyl H3K9 and dimethyl H3K27, Ta3 sequences were not well amplified from ChIP samples using antibodies against trimethyl H3K9 and trimethyl H3K27, consistent with previous reports (Johnson et al. 2004; Mathieu et al. 2005). Thus Ta3 lanes in these cases only serve as background controls.
RNA analysis
Total RNA was isolated using TRI reagent (Sigma-Aldrich) according to the manufacturer’s instructions. Isolated RNA samples were further purified using the RNeasy Mini Kit (Qiagen). First-strand cDNA synthesis was performed on 2 μg of RNA using the M-MLV System for RT–PCR (Promega) followed by PCR amplification with ExTaq DNA Polymerase (Takara Mirus) according to the manufacturers’ recommendations. Primers to amplify FLC, FLM, MAF2∼5, UBIQUITIN, FT, SOC1, TSF, AGL24, and VIN3 were previously described (Ratcliffe et al. 2001, 2003; Michaels et al. 2003; Sung and Amasino 2004). The primer pair used to amplify VIL1 was 5′-CACCTGCAAGAATGCATCGTGTAGAGCTAA TGT-3′ and 5′-TGCCTTTTTAATTGCAGAAGTGCAAAGCT-3′. The primer pair used to amplify VIL2 was 5′-CACCTGCAAAAATTTGGCT TGTCGTGCTGTACT-3′ and 5′-AAGAGATTCAAGAGCAGAAGAG CAAAGTT-3′. The primer pair used to amplify VIL3 was 5′-CACCTGT CAGAACTTGGCTTGCCAGAATAAACT-3′ and 5′-AAGCTCTTTCA AAGCAGAAGAGCAATGTT-3′. Amplified fragments were separated on a 1%–4% agarose gel.
Acknowledgments
We are grateful to Tifani Eshoo for technical assistance, and Scott Woody and Amanda Johnson for their comments on the manuscript. We thank the Salk Institute Genome Analysis Laboratory for providing knockout pools containing alleles of vil1 and flm. We thank Yuehui He for the isolation of flm-3. We also thank Eileen Maher and the Molecular Interaction Facility of the University of Wisconsin Biotechnology Center for performing the yeast two-hybrid screen that identified VIL1. Research in R.M.A.’s laboratory was supported by the College of Agricultural and Life Sciences of the University of Wisconsin and by grants from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program and the National Science Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1493306
References
- Alonso, J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. doi: 10.1038/nature02269. [DOI] [PubMed] [Google Scholar]
- Bienz, M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Martienssen, R., Colot, V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Wood, C.C., Robertson, M., James Peacock, W., Dennis, E.S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006;46:183–192. doi: 10.1111/j.1365-313X.2006.02686.x. [DOI] [PubMed] [Google Scholar]
- Jacobs, S.A., Khorasanizadeh, S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., Dean, C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., Jacobsen, S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Mollah, S., Garcia, B.A., Muratore, T.L., Shabanowitz, J., Hunt, D.F., Jacobsen, S.E. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I., Amasino, R.M. Effect of vernalization, photoperiod, and light quality on the flowering phenotype of Arabidopsis plants containing the FRIGIDA gene. Plant Physiol. 1995;108:157–162. doi: 10.1104/pp.108.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault, M., Tessadori, F., Germann, S., Snijder, B., Fransz, P., Gaudin, V. The Arabidopsis LHP1 protein is a component of euchromatin. Planta. 2005;222:910–925. doi: 10.1007/s00425-005-0129-4. [DOI] [PubMed] [Google Scholar]
- Mathieu, O., Probst, A.V., Paszkowski, J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., Ditta, G., Gustafson-Brown, C., Pelaz, S., Yanofsky, M., Amasino, R.M. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003;33:867–874. doi: 10.1046/j.1365-313x.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., Himelblau, E., Kim, S.Y., Schomburg, F.M., Amasino, R.M. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 2005;137:149–156. doi: 10.1104/pp.104.052811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi, K., Jasencakova, Z., Schubert, I., Goto, K. The Arabidopsis heterochromatin protein1 homolog (TERMINAL FLOWER2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol. 2005;46:1747–1756. doi: 10.1093/pcp/pci195. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Nadzan, G.C., Reuber, T.L., Riechmann, J.L. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001;126:122–132. doi: 10.1104/pp.126.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., Riechmann, J.L. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci, K.C., Michaels, S.D., Amasino, R.M. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Krober, S., Amasino, R.A., Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes & Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., Dennis, E.S. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell. 1999;11:445–458. doi: 10.1105/tpc.11.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S., Amasino, R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- Sung, S., He, Y., Eshoo, T.W., Tamada, Y., Johnson, L., Nakahigashi, K., Goto, K., Jacobsen, S.E., Amasino, R.M. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 2006;38:706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, A., Kobayashi, Y., Goto, K., Abe, M., Araki, T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46:1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]