Abstract
RNA silencing refers to small regulatory RNA-mediated processes that repress endogenous gene expression and defend hosts from offending viruses. As an anti-host defense mechanism, viruses encode suppressors that can block RNA silencing pathways. Cucumber mosaic virus (CMV)-encoded 2b protein was among the first suppressors identified that could inhibit post-transcriptional gene silencing (PTGS), but with little or no effect on miRNA functions. The mechanisms underlying 2b suppression of RNA silencing are unknown. Here, we demonstrate that the CMV 2b protein also interferes with miRNA pathways, eliciting developmental anomalies partially phenocopying ago1 mutant alleles. In contrast to most characterized suppressors, 2b directly interacts with Argonaute1 (AGO1) in vitro and in vivo, and this interaction occurs primarily on one surface of the PAZ-containing module and part of the PIWI-box of AGO1. Consistent with this interaction, 2b specifically inhibits AGO1 cleavage activity in RISC reconstitution assays. In addition, AGO1 recruits virus-derived small interfering RNAs (siRNAs) in vivo, suggesting that AGO1 is a major factor in defense against CMV infection. We conclude that 2b blocks AGO1 cleavage activity to inhibit miRNA pathways, attenuate RNA silencing, and counter host defense. These findings provide insight on the molecular arms race between host antiviral RNA silencing and virus counterdefense.
Keywords: Viral suppressor, Cucumber mosaic virus 2b, RNA silencing, AtAGO1, cleavage activity, counter defense
RNA silencing refers to small regulatory RNA-mediated processes that repress gene expression. A variety of pathways are implicated in RNA silencing, but they all share certain core biochemical features. RNA silencing starts with the processing of double-stranded RNAs (dsRNAs) or imperfectly self-folded hairpin precursors into small interfering RNA (siRNA) or miRNA duplexes, by RNase III-type Dicer enzymes. The small RNA duplexes are then incorporated into a ribonucleoprotein complex called RNA-induced silencing complex (RISC) that degrades any RNA complementary to small RNAs (Bartel 2004; Tomari and Zamore 2005). The core component of the RISC complex is an Argonaute (AGO) protein, which has a PAZ domain that provides a 3′ 2-nucleotide overhang recognition pocket and a PIWI domain that confers endonucleolytic activity (Hall 2005; Song and Joshua-Tor 2006). In Arabidopsis, the AGO family contains 10 putative members (Fagard et al. 2000), among which AGO1 is the best characterized. The involvement of AGO1 in RNA silencing was initially uncovered by genetic analysis because strong ago1 alleles exhibit severe defects in miRNA accumulation, target mRNA cleavage, and resulting developmental anomalies (Vaucheret et al. 2004). Moreover, ago1 mutants are impaired in spontaneous silencing of a transgene (Fagard et al. 2000) and exhibit hypersusceptibility to Cucumber mosaic virus (CMV) (Morel et al. 2002). The biological role of AGO1 in RNA silencing has been confirmed by two recent biochemical studies demonstrating AGO1 cleavage activity in vitro (Baumberger and Baulcombe 2005; Qi et al. 2005).
RNA silencing can be mediated at either the epigenetic or post-transcriptional level in plants. One aspect of post-transcriptional gene silencing (PTGS) reduces endogenous gene expression mainly though miRNA pathways (Jones-Rhoades et al. 2006), which require DCL1 and AGO1 functions in Arabidopsis. Another aspect of PTGS defends hosts against exogenous nucleic acid invaders (i.e., transgenes, viruses) through siRNA pathways, which involve different combinations of RNA-dependent RNA polymerases (RDRs), DCLs, AGOs, and other components (Voinnet 2005). For example, PTGS of a sense transgene (S-PTGS) is triggered by dsRNAs that are converted from single-stranded RNAs (ssRNAs) by RDR6 with the help of SGS3 (Vaucheret 2006). Virus-induced gene silencing (VIGS) is elicited by dsRNA intermediates of replicating viruses, RDR1- or RDR6-mediated formation of dsRNAs, or highly structured regions of viral RNAs (Voinnet 2005). As an adaptive immune system VIGS allows cells to degrade exogenous virus invaders through the action of cis-acting siRNAs that are derived from and target the invaders. In addition, VIGS may also generate a mobile silencing signal that primes RNA silencing in uninfected cells (Baulcombe 2004; Voinnet 2005). Therefore, systemic infection of plants would require effective mechanisms elaborated by the virus to suppress host RNA silencing.
As a counter strategy, many plant and animal viruses have evolved suppressor proteins to block host RNA silencing at various stages of the siRNA pathways and in many cases miRNA pathways as well (Anandalakshmi et al. 1998; Brigneti et al. 1998; Kasschau and Carrington 1998; Chapman et al. 2004; Chen et al. 2004; Dunoyer et al. 2004; Mlotshwa et al. 2005). The suppression of siRNA pathways is accompanied by a significant reduction or complete elimination of siRNAs. Interference with miRNA pathways in Arabidopsis has been associated with developmental defects, which partially phenocopied ago1 mutants and mutants with deficiency in genes involved in miRNA biogenesis and metabolism (dcl1, hen1, and hyl1). The molecular mechanisms of silencing suppression by several suppressors have been investigated, among which the best characterized so far is the tombusviral P19 protein. P19 forms head-to-tail homodimers that sequester siRNA duplexes and prevent them from entering RISC (Vargason et al. 2003; Ye et al. 2003). P21 of Beet yellow virus and P1/HC-Pro of Turnip mosaic virus (TuMV) may function similarly (Ye and Patel 2005; Lakatos et al. 2006). It has also been proposed that the B2 protein of Flock House virus binds dsRNAs and siRNAs and inhibits siRNA formation (Chao et al. 2005), and P38 suppresses DCL4 activities (Deleris et al. 2006).
CMV 2b was among the first two suppressors identified that could inhibit PTGS of sense-GFP transgenes (S-PTGS) (Brigneti et al. 1998), with little or no effect on miRNA functions (Chapman et al. 2004). To define the CMV 2b site of action in RNA silencing, we investigated its suppression mechanism. Here, we showed that CMV 2b also blocks miRNA pathways eliciting developmental anomalies partially phenocopying ago1 mutant alleles. We found that CMV 2b interacts directly with AGO1 and inhibits its cleavage activity in RISC reconstitution assays and that AGO1 recruits virus-derived siRNAs in vivo. These results indicate that CMV 2b blocks AGO1 cleavage activity to attenuate RNA silencing and counteract host defense.
Results
CMV 2b suppressor causes developmental abnormality in Arabidopsis
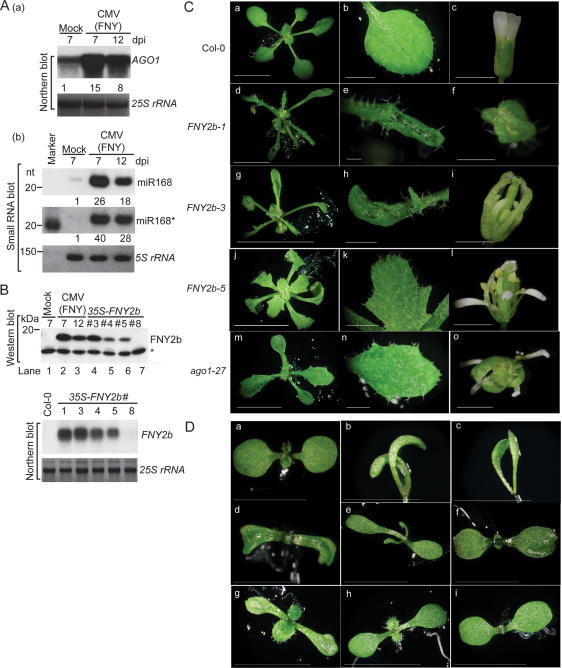
CMV-encoded 2b protein was among the first two suppressors identified that could inhibit PTGS (Brigneti et al. 1998), but with little or no effect on miRNA functions (Chapman et al. 2004). Considering that the 2b suppressor previously examined (Chapman et al. 2004) was from a mild strain (Q strain) of CMV (Guo and Ding 2002), we investigated whether the homologous suppressor from a severe strain (FNY strain) (Rizzo and Palukaitis 1990) might behave differently. CMV (FNY) infection causes severe symptoms in Arabidopsis, including stunting, shortened internodal distances, twisted stems, and small-sized flowers, which were unable to open (data not shown). Figure 1 shows that FNY-infected plants contained abundant 2b protein and elevated levels of miRNAs, as exemplified by miR168, their star strands and target mRNAs (Fig. 1A [panels a, b], B [top panel, lanes 1–3]), indicating inhibition of miRNA pathways.
Figure 1.
CMV 2b causes developmental anomalies in Arabidopsis. (A) CMV (FNY strain) infection enhances accumulation of AGO1 (panel a) and miR168 and miR168* (panel b). Samples were collected 7 or 12 d post-inoculation (dpi). Control plants were treated with buffer only (Mock). The numbers below each lane refer to expression levels relative to WT, after normalization against the loading controls (25S or 5S rRNAs). (B) FNY2b transcript and protein levels in 35S-FNY2b lines and CMV (FNY strain)-infected plants. Polyclonal antibody against FNY2b was used. (Top panels, lanes 1–3) For CMV infection experiment, samples were collected 7 or 12 dpi. Transgenic line numbers are given on top. A cross-reacting band (asterisk) was used as a loading control. (C) Morphological phenotypes of 35S-FNY2b transgenic lines. Photographs were taken for 4-wk-old seedlings, their fourth true leaves, and adult flowers of WT Col-0 (panels a–c) and representative 35S-FNY2b lines (#1, #3, and #5), and for 3-wk-old seedlings, second true leaves, and adult flowers of ago1-27 (panels m–o). (Panels d–f) 35S-FNY2b-1. (Panels g–i) 35S-FNY2b-3. (Panels j–l) 35S-FNY2b-5. Bars: panels a,d,g,j,m, 10 mm; panels b,e,h,k,n, 2 mm; panels c,f,i,l,o, 1 mm. (D) 35S-FNY2b, a series of ago1 mutant alleles and WT Col-0 seedlings at the cotyledon stage. (Panel a) WT Col-0. (Panel b,c) Extremely severe 35S-FNY2b lines with unexpanded cotyledons. (Panels d–f) 35S-FNY-3, 35S-FNY-4 and 35S-FNY-5, respectively. (Panels g–i) ago1-36, ago1-25, and ago1-27. Bars, 3 mm.
Since interference with miRNA pathways is a common feature of many viral suppressors (Mallory et al. 2002; Kasschau et al. 2003; Chapman et al. 2004; Dunoyer et al. 2004; Mlotshwa et al. 2005), we hypothesized that the molecular phenotypes of FNY-infected plants could be attributed to the 2b suppressor itself. To this end, we compared transgenic Arabidopsis plants expressing 35S-CMV 2b transgenes from two CMV strains, FNY and Q. FNY2b and Q2b proteins share 51% identity and 62% similarity in amino acid sequence (Rizzo and Palukaitis 1990; Guo and Ding 2002). Approximately 80% (104 of 132) of 35S-FNY2b primary transformants exhibited developmental abnormalities with varying severity (Fig. 1C). The most severe transgenic lines (e.g., FNY2b-1) (Fig. 1C, panels d–f) had elongated, narrow, serrated, and strongly upward curled leaves. In 20% of the lines, the leaves appeared to have lost their polarity with twisted petioles. Flowers from these plants were very compact with narrow sepals and petals separated by gaps and were mostly sterile. Approximately half of the flowers remained closed, whereas the remaining flowers carried anthers devoid of pollen. Lines with less severe phenotype (e.g., FNY2b-3) displayed serrated, lobed, or curled rosette leaves, and the plants were partially sterile because their flowers contained shorter stamens (Fig. 1C, panels g–i). Lines with mild developmental defects are represented by FNY2b-5 (Fig. 1C, panels j–l). These plants were fertile and had slightly curled or uncurled rosette leaves with strong serration. Their sepals and petals turned around and appeared to have a partial loss of adaxial and abaxial identity; however, their carpels and stamens remained fertile. All these morphological characteristics phenocopied ago1-25 and ago1-27, two weak ago1 mutant alleles (Fig. 1C, panels m–o; Morel et al. 2002; data not shown). Northern and Western blot analyses showed that the phenotypic severity was related to FNY2b expression levels (Fig. 1B). In T2 or subsequent generations, the abnormal phenotypes in 35S-FNY2b lines #3–5 were also easily distinguishable from those of wild type (WT) and of line 35S-FNY2b-8 (Fig. 1D, panels a, d–f). Soon after germination, lines #3–5 seedlings had narrow and curved, or cup-shaped or spoon-shaped, cotyledons with dark-green color phenocopying ago1 mutant alleles (Fig. 1D, panels g–i). Rescreening of 35S-FNY2b primary transformants yielded ∼10% of extremely severe transformants with unexpanded cotyledons (Fig. 1D, panels b,c), mimicking ago1-1 and other severe and nonviable ago1 mutant alleles (Bohmert et al. 1998; Kidner and Martienssen 2004; data not shown). Severe 35S-FNY2b lines died soon after emergence of true leaves.
In contrast to 35S-FNY2b lines, only 0.6% of 35S-Q2b transgenic lines (seven of 1208) displayed obvious developmental anomalies (data not shown). The phenotypic difference elicited by expression of FNY2b and Q2b is likely related to their expression levels. The majority of the 35S-Q2b lines had comparable transcript levels to the 35S-FNY2b lines (Fig. 1B; Supplementary Fig. S1A), but only <5% of the lines contained barely detectable Q2b proteins. Moreover, the majority of the detected Q2b proteins in these lines were truncated in contrast to at least 80% of the 35S-FNY2b lines, which expressed full-length FNY2b at high levels (Fig. 1B; Supplementary Fig. S1A). To avoid a potential difference in the antibody-binding affinity to the two proteins, we investigated 2b protein levels in transgenic plants expressing 35S-FNY2b-3HA and 35S-Q2b-3HA. Q2b-3HA was also barely detectable, and 90% of the protein appeared truncated, while FNY2b-3HA was intact and stable (Supplementary Fig. S1B,C).
Inhibition of miRNA- and siRNA-guided cleavage of mRNA targets
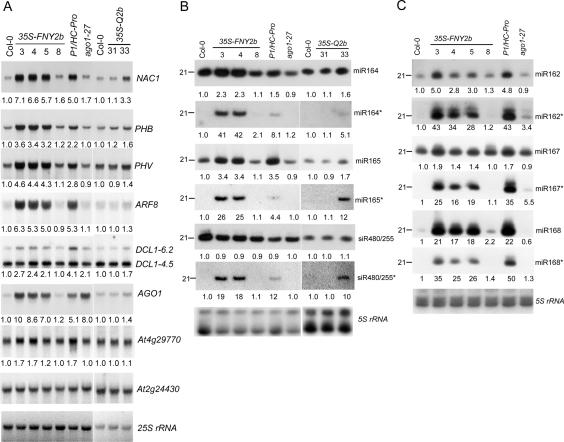
Developmental anomalies in Arabidopsis expressing viral suppressors have been associated with aberrant regulation of miRNA- or siRNA-targeted mRNA expression (Kasschau et al. 2003; Chapman et al. 2004; Dunoyer et al. 2004). We compared the expression level of several miRNA- or siRNA-guided mRNAs between WT and CMV 2b transgenic plants. Plants expressing the P1/HC-Pro-silencing suppressor of TuMV (Mlotshwa et al. 2005) and ago1-27, a weak allele of ago1, were used as controls (Fig. 2A). Previous reports identified NAC1, PHB, PHV, ARF8, and AGO1 as targets of miR164, mir165, miR167, and miR168, respectively (Kidner and Martienssen 2004; Vaucheret et al. 2004; Guo et al. 2005). Figure 2A shows that in all 35S-FNY2b lines with distinguishable phenotypes, the accumulation of these miRNA-guided mRNAs was enhanced several-fold and their expression levels were either similar to or even higher than those in P1/HC-Pro plants. Greater accumulation of NAC1 and AGO1 transcripts was also detected in ago1-27, because of the loss of AGO1 cleavage activity (Vaucheret et al. 2004). DCL1 transcripts were reported to comprise a full-length form (6.2 kb), and two other shorter forms (a 4.5-kb mRNA containing 5′-DCL1 sequence and a 2.5-kb mRNA containing 3′-DCL1 sequence), and only the full-length form is targeted by miR162 (Xie et al. 2003). Figure 2A shows that the full-length DCL1 level was also up-regulated twofold to threefold in 35S-FNY2b plants (lines #3–5) but not in line #8. At2g49770 is guided by endogenous ta-siRNA480/255, a trans-acting siRNA whose production is initiated by molecular phasing cleavage of another miRNA-directed primary transcript of At2g27400 (Allen et al. 2005). Expression of 35S-FNY2b had less effect on At4g29770 than on miRNA targets. Similar analysis of 35S-Q2b #31 and #33 showed that only #33 (with weak developmental anomalies) accumulated more miRNA-guided targets (Fig. 2A). Together, our results suggest that CMV 2b inhibits in vivo endonucleolytic cleavage of mRNAs guided by small RNAs.
Figure 2.
Accumulation of small RNAs and their * strands and target mRNAs in transgenic Arabidopsis plants expressing CMV 2b. (A) Northern blot analysis of high-molecular-weight RNAs. Transgenic line numbers are given on top. Blots were hybridized with the indicated gene-specific probes. A non-small-RNA-targeted mRNA (At2g24430) was used as a loading control. (B,C) Low-molecular-weight RNA blot. Duplicate blots were hybridized to the oligonucleotide probes complementary to the small RNAs, and their star strands are indicated. The numbers below each lane in A–C refer to expression levels relative to WT, after normalization against the loading control. For DCL1 in A, only the full-length transcript (∼6.2 kb) is targeted by miR162, and its accumulation level was calculated and is shown below.
Altered accumulation of small RNAs and small RNA*s
We next investigated expression levels of small RNAs targeting the mRNAs analyzed above (Fig. 2B,C). There was increased accumulation of all tested miRNAs but not ta-siRNA480/255 in 35SS-FNY2b plants (lines #3–5), as was the case with the P1/HC-Pro plants. Among transgenic lines expressing 35S-Q2b, line #33 contained slightly elevated levels of miRNAs (Fig. 2B). In contrast to miRNAs and guide-strand siRNAs, miRNA*s and passenger-strand siRNAs (siRNA*) are normally expressed at or below the detection limit of small RNA blots in WT Arabidopsis. This is presumably because passenger strands of siRNA or miRNA* are preferentially excluded from the RISC complex and rapidly degraded (Jones-Rhoades et al. 2006). Figure 2, B and C, shows enhanced accumulation of all tested miRNA*s in 35S-FNY2b plants and slight accumulation in 35S-Q2b-#33 plants, as found in transgenic plants overexpressing P1/HC-Pro, P19, and P21 (Fig. 2B,C; Chapman et al. 2004; Mlotshwa et al. 2005). Note that the passenger strand of ta-siRNA480/255 accumulated to a greater extent, although its guide strand’s accumulation was slightly decreased. Similar results were obtained from inflorescence tissues (data not shown).
CMV 2b interferes with miRNA pathways by a mechanism different from that used by P19
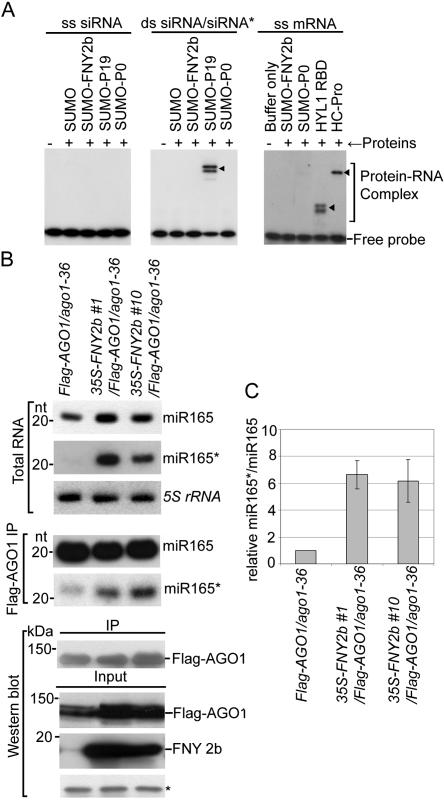
One explanation for the aberrant accumulation of miRNA* in plants expressing silencing suppressors P19, P21, and P1/HC-Pro is that these proteins hijack and stabilize miRNA/miRNA* duplexes, preventing their loading into the RISC complex (Ye et al. 2003; Lakatos et al. 2006). To investigate if CMV 2b shares this strategy, we tested if this suppressor binds small RNAs. Whereas P19 bound small RNA duplexes, purified recombinant FNY2b and Q2b bind neither single-strand (ss) siRNA nor siRNA duplexes in vitro (Fig. 3A, left and middle panels). Similarly, CMV 2b, unlike HYL1 and HC-Pro, did not bind ss mRNA (Fig. 3A, right panel). To further investigate if 2b prevents incorporation of miRNAs into RISC, we created double-transgenic plants expressing 35S-FNY2b in a Flag-AGO1/ago1-36 background (Baumberger and Baulcombe 2005). Figure 3B shows that miRNAs were readily loaded into the RISC complex in FNY2b-expressing plants compared with WT. Moreover, the relative ratio of miR165*/miR165 was significantly higher in Flag-AGO1 immunoprecipitates derived from 2b transgenic plants than from control plants (Fig. 3C). Based on these results, we concluded that CMV 2b suppresses RNA silencing by a mechanism different from that used by P19, P21, and P1/HC-Pro.
Figure 3.
CMV 2b suppresses RNA silencing by a mechanism different from that used by P19. (A) Binding of ss and ds siRNAs and ss mRNA in vitro. Complexes were analyzed using native PAGE. (B) Analysis of miRNA and miRNA* in RISC of 2b-expressing plants. RNA were extracted from flowers or Flag-AGO1 immunoprecipitates of transgenic plants (two independent lines, #1 and #10) harboring 35S-FNY2b in a Flag-AGO1/ago1-36 background (Baumberger and Baulcombe 2005) and Flag-AGO1/ago1-36 control plants (Flag-AGO1/ago1–36). For middle panels, each lane contained small RNAs associated with Flag-AGO1 immunoprecipitated from 0.2 g of flowers. (Bottom panels) The input and immunoprecipitates of Flag-AGO1 and FNY2b were analyzed by Western blot assays in the same samples for small RNA blots. A cross-reacting band (*) was used as a loading control. (C) The relative ratio of miR165* and miR165 is increased in Flag-AGO1 immunoprecipitates from 35S-FNY2b-overexpressing lines compared with those from control plants. The mean signal values of miR165* and miR165 levels in 35S-FNY2b were calculated relative to those in control plants (Flag-AGO1/ago1-36), where the ratio was arbitrarily assigned a value of 1. Standard errors represent at least four experiments.
2b interacts with AGO1 in vivo
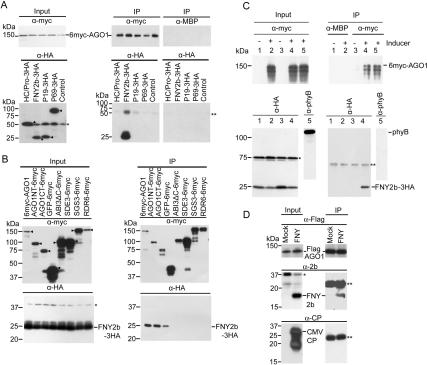
Interference with small RNA-guided mRNA endonucleotic cleavage but not with upstream pathways of small RNA/small RNA* led us to hypothesize that the CMV 2b may inhibit RISC activity. To examine whether there is interaction between CMV 2b and AGO1, we transiently expressed 35S-6myc-AGO1 together with 35S-FNY2b-3HA or other control proteins in Nicotiana benthamiana. Figure 4A shows that antibody to 6myc-AGO1 specifically coimmunoprecipitated FNY2b-3HA, but not with P1/HC-Pro, P19, and P69, all of which were tagged with 3HA at their C termini. Similar coimmunoprecipitation experiments showed that FNY2b-3HA did not interact with 6myc-tagged SDE3, SGS3, and RDR6, all of which play a role in PTGS pathways, nor with two other control proteins, ABI3 (Zhang et al. 2005) and GFP (Fig. 4B). Similar interaction results were obtained with Q2b-3HA. Specific interaction between FNY2b and AGO1 was further confirmed by their colocalization in the nucleus and in cytoplasmic foci when coexpressed in N. benthamiana cells (Supplementary Fig. S2).
Figure 4.
Specific interaction of CMV 2b and AGO1 in vivo. (A,B) Coimmunoprecipitation of FNY2b and AGO1 transiently expressed in N. benthamiana. Proteins were tagged either with 6myc or 3HA. Total crude protein extracts (Input) were immunoprecipitated (IP) with polyclonal antibody to myc or MBP. Western blots were analyzed with a monoclonal antibody to myc to detect myc-tagged proteins (top panels), and a monoclonal antibody to HA to detect coimmunoprecipitated HA-tagged proteins (bottom panels). Arrowheads (◀) indicate transiently expressed proteins. (C) Interaction of FNY2b and AGO1 proteins in Arabidopsis plants. Two-week-old seedlings expressing 35-FNY2b-3HA/XVE-1-6myc-AGO1 were treated overnight with 50 μM MG132 in the absence (lanes1,3) or presence (lanes 2,4,5) of β-estradiol (25 μM) to induce 6myc-AGO1 expression. Total protein extracts were immunoprecipitated (IP) with polyclonal antibody to myc or MBP. Western blots were analyzed with a monoclonal antibody to myc to detect 6myc-AGO1 (top panel), and a monoclonal antibody to HA to detect coimmunoprecipitated FNY2b-3HA (bottom panel). A monoclonal antibody to phyB was used as a negative control to detect the presence of phyB. (D) CMV-encoded 2b suppressor is incorporated into the AGO1 complex in Arabidopsis plants during viral infections. Flag-AGO1/ago1-36 plants at flowering stages were treated with buffer only (mock) or infected with the CMV FNY strain. Total protein extracts from flowers were immunoprecipitated (IP) with monoclonal antibody to Flag. Western blots were analyzed with the same antibody to detect Flag-AGO1 (top panel), and a polyclonal antibody to FNY2b to detect coimmunoprecipitated FNY2b (middle panel). A polyclonal antibody to CMV CP was used as a negative control to detect the presence of coat protein. (A–D) In all panels, an asterisk (*) indicates a cross-reacting band in the input fraction, which served as a negative control since it was absent in the IP fractions. Double asterisks (**) indicate the heavy or light chains of the protein A-conjugated antibody.
Next, we investigated whether FNY2b/AGO1 interaction also occurs in Arabidopsis plants using double-transgenic lines harboring 35S-FNY-3HA and XVE-6myc-AGO1. Upon induced expression of 6myc-AGO1, antibody to 6myc-AGO1 was able to coimmunoprecipitate FNY2b-3HA, but not control proteins, such as phyB and a cross-reacting protein (Fig. 4C). Furthermore, in Flag-AGO1/ago1-36 plants infected with CMV (FNY), the viral 2b (Fig. 4D, middle panel) but not its coat protein (Fig. 4D, bottom panel), was coimmunoprecipitated with Flag-AGO1 (Fig. 4D, top panel), indicating specific FNY2b–AGO1 interaction (Fig. 4D).
CMV 2b directly binds to one surface of the PAZ-containing module of AGO1 in vitro
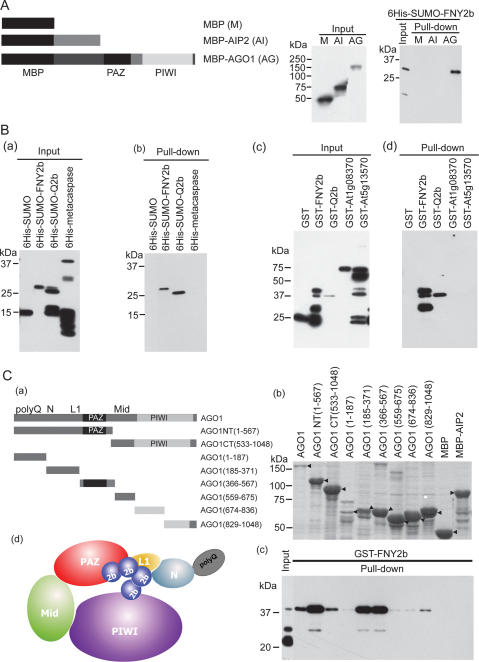
We performed in vitro pull-down assays using various purified proteins to examine if the FNY2b–AGO1 interaction was direct or indirect. Figure 5A shows that maltose-binding protein (MBP)-AGO1, but not MBP and MBP-AIP2 (ABI3-Interacting Protein 2) (Zhang et al. 2005), was able to pull down 6His-SUMO-FNY2b. This interaction was specific as MBP-AGO1 was unable to pull down other control proteins (Fig. 5B). To further define protein domains responsible for the specific interaction, we generated eight truncated AGO1 mutants (Fig. 5C, panels a,b). Pull-down assays showed that an AGO1 NT (amino acids 1–567) truncated mutant interacts with FNY2b at a high affinity, whereas an AGO1 CT (amino acids 533–1048) truncated mutant interacts with FNY2b at a reduced affinity in vitro, consistent with the results of coimmunoprecipitation in vivo (Fig. 4B). Additional pull-down assays showed that FNY2b interacted with three fragments of AGO1—AGO1(185–371), AGO1(366–567), and AGO1(829–1048)—with a stronger affinity to the PAZ domain and the domain similar to the L1 part of Aquifex aeolicus (Aa) and Pyrococcus furiosus (Pf) AGOs (Fig. 5C; Song et al. 2004; Yuan et al. 2005). Sequencing alignment of AtAGO1, AaAGO, and PfAGO, together with structural analyses of AaAGO and PfAGO (Song et al. 2004; Yuan et al. 2005), revealed that AGO1(185–371) and AGO1(366–567) correspond to one surface of the PAZ-containing module of AGOs harboring small RNA and its target mRNA-binding groove, whereas AGO1(829–1048) corresponds to the PIWI-box of AGOs. Collectively, our results indicate that CMV 2b interacts with AGO1 on one surface of the PAZ-containing module harboring the RNA-binding groove and part of the PIWI-box (Fig. 5C, panels c,d).
Figure 5.
CMV 2b directly interacts with AGO1 in vitro. (A) In vitro pull-down assays of 6His-SUMO-FNY2b with three bait proteins: MBP (M), MBP-AIP2 (AI), and MBP-AGO1 (AG). (Left panel) A schematic diagram of the three bait proteins. (Middle panel) The input of the bait proteins. (Right panel) The input and output of 6His-SUMO-FNY2b. Western blots were analyzed with monoclonal antibodies to MBP (middle panel) or to 6His (right panel). (B). In vitro pull-down assays of several 6His- or GST-tagged proteins by MBP-AGO1. (Panels a,b) The input and output of the indicated 6His-tagged target proteins, respectively. (Panels c,d) The input and output fraction of the indicated GST-tagged target proteins, respectively. Western blots were analyzed with monoclonal antibodies to 6His (panels a,b) or to GST (panels c,d). (C) CMV 2b interacts with one face of AGO1. (Panel a) A schematic diagram of full-length and truncated forms of AGO1. The numbers refer to the amino acid residues in the WT AGO1 protein. Locations of the PAZ and PIWI domains are shown. All bait proteins were tagged with MBP at the N terminus. (Panel b) Coomassie brilliant blue G250 staining of the bait proteins, showing their mobility. The major band representing the bait protein is indicated with arrowheads (◀). (Panel c) The output of pulled-down GST-FNY2b. Western blots were analyzed with a monoclonal antibody to GST. All MBP-tagged bait proteins and 6His- and GST-tagged target proteins were purified from Escherichia coli using amylose resins, nitrilotriacetate resins, and glutathione resins, respectively. In all assays, 2 μg of target proteins were pulled down with the indicated bait proteins (2 μg each) using amylose resins. (Panel d) A cartoon showing binding of CMV 2b to one surface of the PAZ-containing module of AGO1, which harbors ss small RNA and its target mRNA-binding groove. The predicted domains of poly-Q (Q), N terminus (N), Linker 1 (L1), Mid, PAZ, and PIWI are shown.
2b suppresses RNA silencing by direct blocking of RISC activity in vitro
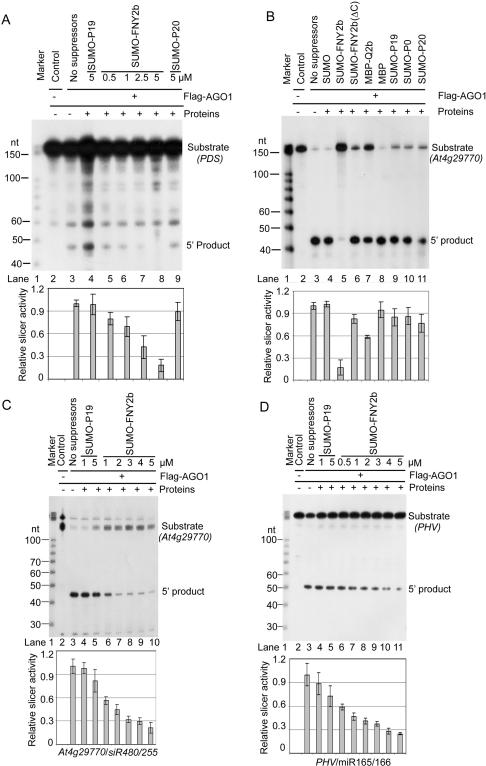
The specific interaction of CMV 2b with AGO1 in vitro and in vivo, the suppression of small RNA-guided target cleavage in vivo, and the increased ratio of miRNA*/miRNA associated with Flag-AGO1 in FNY2b transgenic lines led us to hypothesize that CMV 2b directly blocks AGO1 cleavage activity. To test this hypothesis, we performed in vitro reconstitution of RISC using immunoprecipitated Flag-AGO1, prepared from Flag-AGO1/AGO1-36 transgenic plants (Baumberger and Baulcombe 2005). Ss siRNAs with photoactive residues at either the 5′ or 3′ end could be specifically cross-linked to immunoaffnity-purified Flag-AGO1, indicating that AGO1 could take up exogenous small RNAs (Supplementary Fig. S3; Qi et al. 2005). In vitro loaded ss siRNA targeting PDS was able to direct Flag-AGO1 to mediate PDS mRNA cleavage at the expected position (Fig. 6A, lane 3). Preincubation of SUMO-FNY2b with Flag-AGO1 before loading of the ss siRNA inhibited PDS cleavage, which was correlated with the amount of FNY2b (Fig. 6A, lanes 5–8). In contrast, preincubation of SUMO-P19 and SUMO-P20 (Lu et al. 2004) had no significant effect on Flag-AGO1 cleavage activity. Therefore, the FNY2b suppressor, indeed, blocks RISC activity in vitro.
Figure 6.
CMV 2b inhibits AGO1 slicer activity in RISC reconstituted in vitro. (A) FNY2b blocks the activity of RISC reconstituted in vitro. (Lanes 3–9) Immunoprecipitates (IPs) containing Flag-AGO1 were prepared from inflorescences of Flag-AGO1/AGO1-36 plants using a monoclonal antibody to Flag. (Lane 2) Similar immunoprecipitates prepared from inflorescences of Flag-AGO1/AGO1-36 plants using a monoclonal antibody to 6His were used as negative controls. Immunoprecipitates were incubated with the indicated suppressor protein (in micromolar concentration; lanes 4–9) or without any suppressor protein (lane 3, buffer only, containing the same ionic concentrations) before mixing with the ss siRNA complementary to the target PDS mRNA. Reconstituted RISC was tested for cleavage activity by incubation with a 32P-cap-labeled PDS mRNA. RNAs recovered from the reaction mix were fractionated on 12% denaturing gels. The positions of intact substrates, 5′ cleavage products, and RNA markers are shown. (B) FNY2b inhibits cleavage activity of Flag-AGO1. Immunoprecipitates containing Flag-AGO1 were incubated without (buffer only, lane 3) or with the indicated proteins (lanes 4–11) before addition of 32P-cap-labeled in vitro transcripts of At4g29770. (Top panel, lane 2) Immunoprecipitates of WT Col-0 inflorescences using Flag antibody were used as a negative control. The final concentration of the indicated suppressors and control proteins was 5 μM except MBP and MBP-Q2b, which were 5 μg (∼1.5 and 1 μM, respectively). RNAs recovered from the reaction mix were analyzed as in A. (C,D) Inhibition of AGO1 cleavage activity is proportional to the FNY2b amount. Immunoprecipitates containing Flag-AGO1 were incubated without (buffer only, lane 3) or with the indicated suppressors at different concentrations before mixing with 32P-cap-labeled in vitro transcripts of At4g29770 (C) and PHAVOLUTA (PHV) (D). (Lane 2) Control immunoprecipitates were prepared from WT Col-0 inflorescences using antibody to Flag. RNAs recovered from the reaction mix were analyzed as in A. (A–D) In all the four panels, the experiments were done at least three times. (Lane 3) In each experiment, the cleavage efficiency was normalized to that obtained with the buffer only. (Bottom panels) Mean values of the relative cleavage efficiency are shown along with standard error.
The interference of FNY2b with RISC reconstitution could be due to either a block of ss siRNA loading into RISC or an inhibition of the intrinsic cleavage activity of AGO1. To further address the mode of FNY2b action, we made use of small RNAs resident in AGO1 in vivo. We confirmed that ta-siR480/255 and miR165/166 could be coimmunoprecipitated with AGO1 (Fig. 3; Supplementary Fig. S3; Baumberger and Baulcombe 2005; Qi et al. 2005). Moreover, the Flag-AGO1-bound siR480/255 could direct At4g29770 transcript cleavage (Fig. 6B [lane 3], C [lane 3]). Figure 6B shows that addition of FNY2b reduced AGO1 cleavage activity at least fivefold, whereas other proteins, SUMO, SUMO-P19, and SUMO-P20, had no significant effect. Another suppressor SUMO-P0 (Pazhouhandeh et al. 2006) also had negligible effect, although it interacted with AGO1 in vitro (data not shown). These results show that FNY2b, indeed, directly blocks AGO1 activity. Interestingly, a truncated mutant of FNY2b, which contains only the N-terminal 50 amino acid residues, could only slightly suppress AGO1-mediated cleavage, suggesting the requirement of intact 2b for full function. Furthermore, MBP-Q2b (∼1 μM) reduced AGO1 slicer activity by approximately twofold compared with MBP alone, suggesting that intact Q2b maintains its suppressing function in vitro (Fig. 6B).
The inhibition of FNY2b on AGO1 slicer activity was concentration-dependent (Fig. 6C). At low FNY2b concentrations, the effect was enhanced by prolonged incubation of the suppressor and AGO1 (Supplementary Fig. S4). Similar results were obtained with miR165/166-mediated cleavage of PHV transcripts (Fig. 6D). Taken together, our results provide evidence that CMV 2b interferes with RNA silencing by directly blocking AGO1 slicer activity.
CMV 2b inhibition on AGO1 activity is a novel mechanism to counter host defense
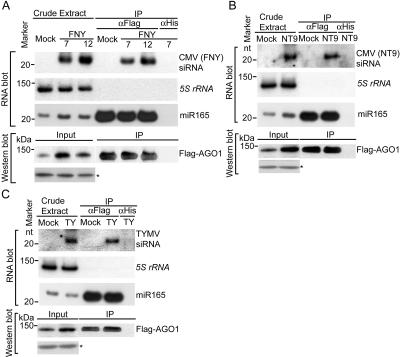
RISC slicer activity is an essential step in VIGS to defend against offending viruses (Deleris et al. 2006). The specific 2b/AGO1 interaction and the inhibition of AGO1 activity in vivo suggest that AGO1 is a major determinant in VIGS. However, a previous study failed to detect viral siRNAs in association with AGO1 immunoprecipitates (Baumberger and Baulcombe 2005). We re-examined this issue and detected viral siRNAs, derived either from CMV (FNY and NT9 strains) or from Turnip yellow mosaic virus (TYMV), in Flag-AGO1 immunoprecipitates recovered from virus-infected plants (Fig. 7A–C). These findings clearly implicate AGO1 function in VIGS. Considering that ago1 mutants are impaired in spontaneous silencing of a transgene (Fagard et al. 2000) and exhibit hypersusceptibility to CMV (Morel et al. 2002), we believe that AGO1 is a major factor directly involved in VIGS and that 2b inhibition of AGO1 activity represents a viral counter strategy to suppress host antiviral silencing.
Figure 7.
AGO1 recruits viral siRNAs. Flag-AGO1/ago1-36 plants were infected with the CMV (FNY strain) (A), the CMV (NT9 strain) (B), and TYMV (C), or with buffer (Mock). Inflorescence tissues were collected 7 dpi for immunoprecipitation with Flag or His antibody except for CMV (FNY) experiments in which samples were harvested at 7 and 12 dpi. Small RNAs were extracted directly from flowers (crude extract) or from Flag-AGO1 immunoprecipitates and control immunoprecipitates obtained with an antibody to 6His (IP). Viral siRNAs were detected by hybridization with gene-specific probes (Materials and Methods). The same membrane was hybridized to a 5S rRNA probe as a control, indicating no small RNA contamination in immunoprecipitate fractions. miR165 was used as a positive control of AGO1–small RNA coimmunoprecipitation. For the crude extract, each lane contained 5 μg of total RNA. The immunoprecipitate samples were derived from 0.3 g of flowers. Two-thirds of the samples were used for siRNA recovery and the remainder for Western blots to monitor Flag-AGO1 levels. Western blot analyses were performed using antibodies to Flag. A cross-reacting band (*) served as a loading control.
Discussion
CMV 2b interferes with miRNA pathways in Arabidopsis
CMV 2b was among the first suppressors identified that could abrogate PTGS of sense-GFP transgene expression in N. benthamiana (Beclin et al. 1998; Brigneti et al. 1998). More recently, CMV 2b from a mild strain (Q) was reported to have very little effect on miRNA-guided functions (Chapman et al. 2004). Here, we showed that CMV 2b from a severe strain (FNY) also interferes with microRNA pathways in Arabidopsis. Several lines of evidence support this claim: (1) The suppressor caused significant miRNA accumulation (Fig. 2B,C). (2) It increased the amount of miRNA* strands and ta-siRNA passenger strands (Fig. 2B,C). (3) It blocked small RNA-guided regulation of mRNAs in vivo (Fig. 2A). (4) Plants expressing 2b displayed obvious developmental defects (Fig. 1C,D) that are strikingly similar to those obtained with other viral suppressors such as P1/HC-Pro, P21, and P19. All these suppressors have been shown to suppress miRNA pathways (Chapman et al. 2004; Dunoyer et al. 2004). Despite phenotypic differences among the transgenic plants, the severity of morphological alterations was generally correlated with the perturbation of the miRNA pathways as determined by miRNA, miRNA*, and target mRNA levels. Notably, many of the defects in FNY2b-overexpressing lines were reminiscent of those observed in ago1 mutant alleles; for example, cup-shaped or spoon-shaped cotyledons, altered shoot meristems, narrow and serrated leaves, and flowers with narrow sepals and petals with gaps (Bohmert et al. 1998; Morel et al. 2002; Baumberger and Baulcombe 2005; Kidner and Martienssen 2005). Therefore, it is reasonable to suggest that most of the observed defects can be attributed to inhibition of miRNA pathways.
In contrast to the FNY2b, the 2b suppressor from the CMV Q strain did not significantly affect miRNA pathways. Comparative analyses clearly demonstrated that Q2b transgenic lines expressed 2b transcript levels as high as FNY2b transgenic lines. This notwithstanding, the Q2b protein was barely detectable in contrast to FNY2b, which accumulated to high levels, indicating differential protein stability. Therefore, the absence of developmental defects in Q2b plants seen here and also reported previously (Chapman et al. 2004) can simply be explained by their low accumulated levels of intact protein.
Results from previous work (Chapman et al. 2004; Dunoyer et al. 2004; Mlotshwa et al. 2005) and this study raise the question whether there are any advantages for virus suppressors to block miRNA pathways. A trivial explanation is that because of shared components, interference in the miRNA pathway by a virus suppressor may be a collateral consequence of its primary effect on siRNA pathways. On the other hand, inhibition of miRNA pathways may be a deliberate strategy for viruses to redirect host gene expression to create a favorable cellular niche for its proliferation. There are two recent reports of cellular miRNAs being involved in host defense (Lecellier et al. 2005; Navarro et al. 2006).
CMV 2b suppresses RNA silencing by direct perturbation of AGO1 slicer activity
Interference with RNA silencing by viral suppressors represents an anti-host defense response by the pathogen. A group of silencing suppressors exemplified by P19 inhibit gene silencing by sequestrating siRNAs and preventing them from entering the RISC (Ye et al. 2003; Lakatos et al. 2006). Our results show that CMV 2b executes its suppressor function not by sequestering siRNAs but by interacting directly with AGO1 and blocking its slicer activity. Several lines of evidence support this notion: (1) 2b and AGO1 colocalized in N. benthamiana cells (Supplementary Fig. S2). (2) 2b and AGO1 specifically associated with each other in vivo in transient expression assays, in transgenic Arabidopsis plants and in CMV-infected plants (Fig. 4A–D). (3) The 2b/AGO1 interaction was specific and direct as shown by in vitro pull-down experiments (Fig. 5A,B). Moreover, we have mapped the primary interaction region on AGO1 to one surface of the PAZ-containing module harboring the RNA-binding groove and part of the PIWI-box (Fig. 5C; Song et al. 2004; Yuan et al. 2005). Finally, and most important, our in vitro RISC reconstitution assays established that 2b inhibits RNA silencing by directly blocking AGO1 activity (Fig. 6; Supplementary Fig. S4).
Our conclusion is consistent with the molecular phenotypes of 2b-overexpressing plants that accumulated mRNA targets because their cleavage by AGO1 was blocked (Figs. 2A, 6). An inhibition of AGO1 slicer activity could similarly lead to elevated levels of the passenger strands of siRNA and miRNA* in our experiments. In Drosophila, siRNA duplexes are loaded into the RISC such that the guide strand of siRNA directs Argonaute-catalyzed cleavage of the passenger strand. Cleavage of the passenger strand facilitates its clearance, liberating the guide strand to pair with target transcripts and direct their cleavage (Matranga et al. 2005; Rand et al. 2005). Although the RISC loading process has not yet been investigated in Arabidopsis, presumably siRNA duplexes share the same properties. If so, 2b likely inhibits cleavage of siRNA passenger strands as well. This inhibition not only blocks clearance of the passenger strands, thereby stabilizing them, but also prevents the guide strand from accessing their mRNA targets, leading to an accumulation of the latter. It was suggested that miRNA/miRNA* duplexes might be loaded into AGO1 by a passenger-strand cleavage-assisted mechanism in Arabidopsis (Matranga et al. 2005). If this is the case, inhibition of AGO1 slicer activity on the miRNA* would also stabilize these star strands and increase the miRNA*/miRNA ratio in AGO1 derived from 2b-overexpressing plants (Figs. 2B,C, 3C).
Two miRNAs, miR162 and 168, negatively regulate the miRNA pathway itself by targeting DCL1 and AGO1 mRNAs, respectively (Xie et al. 2003; Vaucheret et al. 2004). Such feedback regulation of two core factors in the miRNA pathway is important to achieve an appropriate balance of miRNA steady-state levels, which depends on miRNA production by DCL1, and miRNA stabilization by AGO1 (Vaucheret 2006; Vaucheret et al. 2006). In our study, CMV 2b suppressed miR162-guided cleavage of DCL1 in 2b-overexpressing plants, resulting in higher DCL1 mRNA levels, which, in turn, promoted additional maturation of miRNAs from pre-miRNAs. Meanwhile, CMV 2b inhibited miR168-guided degradation of AGO1 mRNA allows more miRNA accumulation. This would explain why the overall miRNA content was increased in 2b-overexpession lines. On the other hand, levels of ta-siRNA480/255, a trans-acting siRNAs, were decreased somewhat in 2b-overexpressing lines. We note that trans-siRNAs are derived from TAS transcripts. TAS primary transcripts, which contain a sequence targeted by specific miRNAs, are first cleaved by AGO1. Through the action of RDR6 and SGS3, the resulting ss TAS transcripts are then converted into dsRNAs, which are subsequently processed into mature ta-siRNAs by DCL4 (Vazquez et al. 2004; Allen et al. 2005). Therefore, upstream inhibition of miRNA-directed phasing by 2b would decrease downstream accumulation of mature ta-siRNAs (Fig. 2B,C). The overall net effect is a slight increase in the transcript levels of At4g29770, which is the target of ta-siR480/255 (Fig. 2A).
A novel strategy to counter host defense
Although RISC slicer activity is a key step in VIGS and defends plants against invading viruses (Deleris et al. 2006), it remains unclear which AGO(s) are involved in this event considering that Arabidopsis contains at least 10 AGO members. AGO1 could be a contributor toward VIGS as it is a key factor in RNA silencing pathways. The role of AGO1 in VIGS is also supported by the observation that ago1 mutants are impaired in spontaneous silencing of a transgene (Fagard et al. 2000) and that these mutants exhibit hypersusceptibility to CMV (Morel et al. 2002). On the other hand, recent work failed to detect siRNAs in association with AGO1 recovered by immunoprecipitation (Baumberger and Baulcombe 2005), a finding inconsistent with the proposed role of AGO1. Here, we demonstrated that AGO1, indeed, recruits viral siRNAs in plants infected with three different viruses. Our biochemical results reported here along with previous genetic evidence (Morel et al. 2002) support the notion that AGO1 is, indeed, a major determinant for VIGS.
Suppressor stability and virus virulence
The 2b suppressor is a pathogenicity determinant of CMV (Brigneti et al. 1998). Our results here showed that the difference in pathogenicity between the two CMV strains is likely due to differential stability of the two suppressors. When expressed in transgenic Arabidopsis, Q2b accumulation was barely detectable, and when detected, the majority of the Q2b protein appeared as truncated forms, suggesting the action of host-specific proteases and/or ubiquitin-mediated processes. This result suggests that host defense against virus infection can be executed at both the protein and the RNA level. In addition to degrading viral RNAs by PTGS, plants can also directly mount a counterattack by destroying virus suppressors using cellular proteases and/or ubiquitin-related processes (Supplementary Fig. S1; data not shown). As a counter host defense mechanism, a virus can undergo mutations in its sequences to stabilize its suppressor, which is apparently the case with the FNY2b.
Conclusions
Our results here support the notion that CMV 2b interferes with miRNA pathways and abrogates RNA silencing by direct interaction with AGO1 and inhibition of its slicer activity. The degree of this inhibition is positively correlated with the protein level. Whether CMV 2b causes any developmental defects depends on the steady level of intact 2b protein in vivo. We have provided evidence that AGO1 is a key factor for VIGS. Direct interference with or usurpation of this core component is an effective strategy for CMV to block VIGS and to evade host defense; we expect this strategy to be used by other suppressors as well. Whether 2b might alter AGO1 conformation upon interaction awaits future crystal structure analysis. We note, however, that Arabidopsis has 10 AGO family members. Similar to the functional redundancy reported for DCLs (Deleris et al. 2006), one or more AGOs may be involved in VIGS-related events. Nevertheless, given the functional conservation, we expect them to share the same or similar biochemical mechanism as AGO1. Therefore, the mode of action of 2b interfering with AGO1 should reflect the mode of 2b inhibition of VIGS. In addition, the 2b suppressor has been previously reported to interfere with movement of a mobile signal of gene silencing (Guo and Ding 2002). Further exploration of other potential functions of CMV 2b will help advance our insight into the regulation of RNA silencing by versatile viral suppressors.
Materials and methods
DNA construction
The majority of constructs were made using the Gateway system (Invitrogen) (Zhang et al. 2005). Several destination vectors (DC in our nomenclature) were created for transient expression in N. benthamiania and stable Arabidopsis transformation. The binary vector pBA002 (Zhang et al. 2005) for constitutive expression in plants was modified to obtain pHyg-DC, pBA-DC, pBA-6myc-DC, and pBA-DC-CFP. The vectors pER8 and pER10 (Zuo et al. 2000) for estradiol-inducible expression in plants under the control of the XVE system were modified to obtain pER8-6myc-DC and pER10-YFP-DC, respectively. Most of the cDNA or DNA fragments were cloned into pENTR/D vectors and then transferred to the appropriate vectors by recombination using the LR Clonase enzyme (Invitrogen). More information is provided in the Supplemental Material.
Transgenic plants
Arabidopsis thaliana (Col-0) plants were transformed with binary vectors pBAFNY2b and pBAQ2b by the floral-dip method (Clough and Bent 1998; Zhang et al. 2006). In some constructs, an epitope tag of 3HA was appended to the C terminus of FNY2b and Q2b. Double-transgenic plants were obtained by the same method by coinfiltrating WT Col-0 plants with two constructs (pBA-FNY2b-3HA/pER8-6myc-AGO1). The Flag-AGO1/ago1-36 line (Baumberger and Baulcombe 2005; Qi et al. 2005) was transformed with pHyg-FNY2b to generate 35S-FNY2b/Flag-AGO1/ago1-36 double transformants. Seeds from infiltrated plants were selected on standard MS medium (Murashige and Skoog 1962) containing the appropriate selective agents: 10 mg/L gluphosinate ammonium (Crescent Chemical Co.) and/or 25 mg/L hygromycin (Sigma) together with 100 mg/L cefotaxime (Sigma).
Expression and purification of recombinant proteins
All recombinant proteins were prepared as previously described (Zhang et al. 2005). MBP fusion proteins included MBP, MBP-AIP2, MBP-AGO1, and serial MBP-truncated AGO1, as well as MBP-HC-Pro. 6His or 6His-SUMO fusion proteins included 6His-metacaspase, 6His-HYL1 (RBD, RNA-binding domain), and 6His-SUMO-(FNY2b, Q2b, P0, P19, P20). The SUMO tag was used to increase the solubility of the recombinant proteins. GST-tagged proteins, including GST-FNY2b, GST-Q2b, GST-At1g08370, and At5g13570, were purified using affinity columns followed by size-exclusion columns. Proteins used for in vitro RISC reconstitution assays were further purified as described (Yuan et al. 2005).
In vitro pull-down assays and coimmunoprecipitation experiments
In vitro pull-down assays and in vivo coimmunoprecipitation were done as described (Zhang et al. 2005). More information is provided in the Supplemental Material.
RNA blot and Western blot analyses
Total RNA was extracted using Trizol reagent from either adult rosette leaves or 2-wk-old seedlings of independent transgenic lines. For Northern blot analyses of high-molecular-weight RNA, hybridizations were performed using indicated gene-specific 32P-radiolabled probes that were synthesized by random priming of cloned cDNA sequences. RNA blot hybridizations of low-molecular-weight RNAs were performed as described (Wang et al. 2004). Blots were hybridized to the 32P-radiolabled oligonucleotide probes complementary to the small RNAs, and their star strands were indicated. Sequences for miRNA* and passenger strands of siRNA were predicted from miRNA precursors or TAS transcript structures (Reinhart et al. 2002; Allen et al. 2005). For blot analyses of both high- and low-molecular-weight RNAs, each lane normally contained 5 μg of total RNA except in Figure 2, where 10 μg were used. 25S rRNA and 5S rRNA (or tRNA) were used as loading controls. Accumulation of small RNA and small RNA* as well as small RNA targets was normalized with respect to loading controls, and relative ratios were calculated relative to WT (Col-0). For analysis of viral siRNAs, flowering Flag-AGO1/ago1-36 plants were infected by sap rub-inoculation with CMV strains (FNY and NT9) and TYMV. Inflorescences tissues were collected 7 and 12 d post-infection for immunoprecipitation for protein analysis and siRNA detection as described below. The pooled probes used for detection of viral siRNAs were synthesized by random priming of cloned virus-specific fragments. For CMV (FNY strain), the primer pairs are FNYRNA1–1196For, 5′-CGTGAGACT GAAGAGATTGCTATGAGG-3′, and FNYRNA1–1820Rev, 5′-GGAGTCGAGTTCCGAAACCTC-3′; FNYRNA2–1029For, 5′-CTGGATCATGTTAGACTTCGACAGTC-3′, and FNYRNA2– 1584Rev, 5′-GATGAAATCTTACCAACAGGAAG-3′; FNYRNA3–1234For, 5′-GTGTCGTAGAATTGAGTCGAGT CATG-3′, and FNYRNA3–1889Rev, 5′-GAATGCGTTGGT GCTCGATGTC-3′. For CMV (NT9 strain), the primer pairs are NT9RNA1–127For, 5′-GCTACTGAGTGTGACCTAGGCC-3′, and RNA1–526Rev, 5′-TAGCTGTGGTGCCGGTATACCC-3′; NT9RNA2For, 5-CACCATGGAATTGAACGTAAGCGCAG-3′, and NT9RNA2Rev, 5′-TCATCCAAACGACCCTTCGGCCC-3′; NT9RNA3For, 5′-CTTGTCGCAACAGCTTTCGCGAC-3′, and NT9RNA3Rev, 5-AGCGCGCATCGCCGAAAGATC-3′. For TYMV, the primer pairs are 1650For, 5′-CTAGTCCT CCTCGCTCTGTCGG-3′, and 2159Rev, 5′-GGTGGGTTCG AAATCGGGTGTGG-3′; 2923For, 5′-GCCAACGAACCGGAC CCACCC-3′, and 3381Rev, 5′-GCAGCCTGAGAGTTTCG GAGGG-3′; 5717For, 5′-GGCCCCTCACCTTTCACCATC-3′, and 6066Rev, 5′-ACCCGGGGGTTCATCATTTCAAG-3′.
Polyclonal anti-CMV CP, FNY2b, and Q2b anti-sera were generated in rabbits immunized with the recombinant full-length CP, FNY2b, and Q2b proteins fused to a 6His tag at their N termini. These antibodies and commercial monoclonal antibodies (α-His [Sigma]; α-myc, -HA, -MBP, -GST [Santa Cruz Biotechnology]) were used for Western blot analyses as indicated in the text.
siRNA preparation and electrophoretic mobility shift assays (EMSAs)
siRNAPDS (5′-UUGUACUGCCAUUAUUCAGdTdT-3′), siRNAPDS* (5′-CUGAAUAAUGGCAGUACAAUU-3′) (IDT), and siRNAluc (5′-IUCGAAGUACUCAGCGUAAGUU-3′) (Dharmacon) were phosphorylated using T4 polynucleotide kinase (PNK, NEB). siRNAs were 5′-end-labeled with either ATP or [γ-32P]ATP depending on the experimental purposes described in the context. Briefly, 10 μM ss siRNA was incubated in a 10 μL of reaction volume containing 1× PNK buffer and 10 U of PNK (10 U/μL) containing either 1 mM ATP or 5 μL (50 μCi) of [γ-32P]ATP (NEN, EASY TIDE). The phosphorylation reactions were incubated for 1 h at 37°C. ss siRNAs were further purified using the Quick Spin oligonucleotide Column (Roche). siRNA duplexes were annealed and purified as described (Rand et al. 2005). EMSAs were done as follows: 1 μM suppressors or control proteins and 1 nM either ss siRNA or siRNA duplex or ss mRNA (At4g29770) were incubated in 15 μL of binding buffer (0.1 M KCl, 25 mM HEPES, 10 mM DTT at pH 7.5) for 15 min at room temperature. The mixture was then separated on a 15% or 6% native gel (Ye and Patel 2005).
Immunoprecipitation of Flag-AGO1-associated small RNAs
Immunoprecipitation of Flag-AGO1-associated small RNAs was performed as described (Chapman et al. 2004). RNA was recovered with Trizol reagent from the immunoprecipitates. The RNA was separated on 8 M urea/15% polyacrylamide gels and subjected to RNA blot analysis of low-molecular-weight RNAs.
Preparation of 32P-cap-labeled in vitro transcripts
Target mRNAs of PDS, PHV, and At4g29770 were synthesized using an in vitro transcription kit (MegaScript; Ambion) from their PCR templates produced from the corresponding pENTR vectors. The mRNA was purified with the phenol/chloroform/isoamyl alcohol method and precipitated with isopropanol following the manufacturer’s instructions. The mRNA was then resuspended in a stock concentration to 10 pmol/μL. The mRNA was then G-capped with [α-32P]GTP (3000 Ci/mmol; NEN) using guanylyl transferase (Ambion). The reaction was performed in 30 μL of volume using 20 pmol of mRNA for 2 h at 37°C. The reaction mix was first passed through a Quick Spin RNA Column (Roche) to remove unincorporated nucleotides. The product was then denatured and run on a 6% urea-acrylamide gel for 30 min at 300 V and then eluted from the gel using Eluta Tube (#k0532; Fermentas). Further purification was done in an identical manner as that for the initial in vitro transcription step. The labeled mRNA was resuspended in the same volume as the original reaction mix. One microliter per reaction was typical for RISC assays.
In vitro reconstitution of RISC
Flag-AGO1 immunoprecipitation and the RISC assay were modified from a previous report (Baumberger and Baulcombe 2005). Briefly, inflorescences of transgenic Flag-AGO1 plants were ground in liquid nitrogen, and proteins were extracted in 4 mL/g fresh material of extraction buffer (20 mM Tris-HCl at pH 7.5, 300 mM NaCl, 5 mM MgCl2, 5 mM DTT, 2× EDTA-free protease inhibitor cocktail [Roche]). Insoluble material was removed by centrifuging twice at 16,000g for 10 min at 4°C. Immunoprecipitation was performed for 2 h at 4°C with 15 μL of α-Flag M2 agarose beads (Sigma) or control antibody (α-His) per 0.2 g of starting materials. The α-Flag M2 agarose beads were washed twice in 1 mL of RISC buffer [40 mM HEPES at pH 7.4, 100 mM KOAc, 5 mM Mg(OAc)2, 4 mM DTT], and Flag-AGO1 was eluted in RISC buffer with 100 μg/mL 3× Flag peptide (Sigma) for 1 h at 4°C (16 μL for 0.2 g of starting material). Sixteen microliters of Flag-AGO1 immunoprecipitate were incubated with 1 μL of the indicated suppressor or control proteins for 1 h at 4°C except in Supplementary Figure S4, where overnight incubation was performed. For controls without any proteins, 1 μL of buffer was added instead. Then, 2 μL of siRNAPDS (1 μM) and 0.3 μL of RNasin were added to the reaction, and incubated for 30 min at room temperature before addition of 3.2 μL of substrate cocktail (1 μL of 20 mM ATP, 1 μL of 32P-labeled and capped mRNA, 1.2 μL of RNase inhibitor). For inhibition with endogenous small RNA-guided cleavage, 3.5 μL of substrate cocktail (1μL of 20 mM ATP, 1 μL of 32P-labeled and capped mRNA, 1.5 μL of RNase inhibitor) were immediately added after 1 h of incubation of Flag-AGO1 and suppressors/proteins. After 2 h at 25°C, RNA was recovered with 250 μL of Trizol reagent according to the manufacturer’s instructions. The labeled RNA was separated on a 12% urea-polyacrylamide gel and detected after exposure to a PhosphorImager plate (Molecular Dynamics).
Acknowledgments
We are grateful to Catherine Bellini for the AGO1 cDNA, Herve Vaucheret for the Arabidopsis L1 line and several ago1 mutant alleles, Vicki Vance for HC-Pro plants, David Baulcombe for Flag-AGO1/ago1-36 lines, Karen Bohmert for ago1-1, Shou-Wei Ding for CMV2b (Q) and Rong-Xiang Fang for CMV2b (FNY) plasmids (originally from Peter Palukaitis), and Keith Perry and Shyi-Dong Yeh for CMV (FNY strain) and CMV (NT9 strain). We also thank Drs. Enno Krebbers, Richard Broglie, and Barbara Mazur for stimulating discussions and critical comments; Erik Nyström and Ronghui Wang for technique assistance; and Jun Xu, Rossana Henriques, Rafael Catala-Rodriguez, and Hui-Wen Wu for cartoon drawing and careful proofreading. The work was supported by a grant from DuPont to N.-H.C.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1495506
References
- Allen, E., Xie, Z.X., Gustafson, A., Carrington, J. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R., Pruss, G., Ge, X., Marathe, R., Mallory, A., Smith, T., Vance, V. A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Baumberger, N., Baulcombe, D. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits miRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beclin, C., Berthome, R., Palauqui, J., Tepfer, M., Vaucheret, H. Infection of tobacco or Arabidopsis by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology. 1998;252:313–317. doi: 10.1006/viro.1998.9457. [DOI] [PubMed] [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., Benning, C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti, G., Voinnet, O., Li, W.X., Ji, L., Ding, S., Baulcombe, D. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chao, J.A., Lee, J.H., Chapados, B.R., Debler, E.W., Schneemann, A., Williamson, J.R. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chapman, E., Prokhnevsky, A., Gopinath, K., Dolja, V., Carrington, J. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes & Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Li, W., Xie, D., Peng, J., Ding, S. Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expression. Plant Cell. 2004;16:1302–1313. doi: 10.1105/tpc.018986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S., Bent, A. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Deleris, A., Gallego-Bartolome, J., Bao, J., Kasschau, K., Carrington, J., Voinnet, O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Lecellier, C., Parizotto, E., Himber, C., Voinnet, O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fagard, M., Boutet, S., Morel, J., Bellini, C., Vaucheret, H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Ding, S.W. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 2002;21:398–407. doi: 10.1093/emboj/21.3.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Xie, Q., Fei, J., Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, T. Structure and function of Argonaute proteins. Structure. 2005;13:1403–1408. doi: 10.1016/j.str.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades, M., Bartel, D., Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kasschau, K., Carrington, J. A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- Kasschau, K., Xie, Z., Allen, E., Llave, C., Chapman, E., Krizan, K., Carrington, J. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kidner, C., Martienssen, R. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Kidner, C., Martienssen, R. The role of ARGONAUTE1 (AGO1) in meristem formation and identity. Dev. Biol. 2005;280:504–517. doi: 10.1016/j.ydbio.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Lakatos, L., Csorba, T., Pantaleo, V., Chapman, E., Carrington, J., Liu, Y., Dolja, V., Calvino, L., Lopez-Moya, J., Burgyan, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier, C., Dunoyer, P., Arar, K., Lehmann-Che, J., Eyquem, S., Himber, C., Saib, A., Voinnet, O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Lu, R., Folimonov, A., Shintaku, M., Li, W.X., Falk, B.W., Dawson, W.O., Ding, S.W. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc. Natl. Acad. Sci. 2004;101:15742–15747. doi: 10.1073/pnas.0404940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A., Reinhart, B., Bartel, D., Vance, V., Bowman, L. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. 2002;99:15228–15233. doi: 10.1073/pnas.232434999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga, C., Tomari, Y., Shin, C., Bartel, D., Zamore, P. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Mlotshwa, S., Schauer, S., Smith, T., Mallory, A., Herr, J., Roth, B., Merchant, D., Ray, A., Bowman, L., Vance, V. Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell. 2005;17:2873–2885. doi: 10.1105/tpc.105.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, J., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., Vaucheret, H. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. doi: 10.1105/tpc.010358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473. [Google Scholar]
- Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Pazhouhandeh, M., Dieterle, M., Marrocco, K., Lechner, E., Berry, B., Brault, V., Hemmer, O., Kretsch, T., Richards, K., Genschik, P., et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. 2006;103:1994–1999. doi: 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, Y., Denli, A., Hannon, G. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Rand, T., Petersen, S., Du, F., Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Reinhart, B., Weinstein, E., Rhoades, M., Bartel, B., Bartel, D. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo, T., Palukaitis, P. Construction of full-length cDNA clones of Cucumber mosaic virus RNAs 1, 2 and 3: Generation of infectious RNA transcripts. Mol. Gen. Genet. 1990;222:249–256. doi: 10.1007/BF00633825. [DOI] [PubMed] [Google Scholar]
- Song, J., Joshua-Tor, L. Argonaute and RNA—Getting into the groove. Curr. Opin. Struct. Biol. 2006;16:5–11. doi: 10.1016/j.sbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Song, J., Smith, S., Hannon, G., Joshua-Tor, L. Crystal structure of argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Zamore, P. Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vargason, J., Szittya, G., Burgyan, J., Hall, T. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes & Dev. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., Bartel, D. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Mallory, A., Bartel, D. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A., Hilbert, J., Bartel, D., Crete, P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Wang, X., Reyes, J., Chua, N.H., Gaasterland, T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z.X., Kasschau, K., Carrington, J. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- Ye, K., Patel, D. RNA silencing suppressor p21 of beet yellows virus forms an RNA binding octameric ring structure. Structure. 2005;13:1375–1384. doi: 10.1016/j.str.2005.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, K., Malinina, L., Patel, D. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y., Pei, Y., Ma, J., Kuryavyi, V., Zhadina, M., Meister, G., Chen, H., Dauter, Z., Tuschl, T., Patel, D. Crystal structure of A-aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Garreton, V., Chua, N.H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes & Dev. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Henriques, R., Lin, S.S., Niu, Q.W., Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral-dip method. Nature Protocols. 2006;1:1–6. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., Chua, N.H. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 2000;24:265–273. doi: 10.1046/j.1365-313x.2000.00868.x. [DOI] [PubMed] [Google Scholar]