Abstract
Neurofibromatosis type 1 (NF1) is among the most common genetic disorders of humans and is caused by loss of neurofibromin, a large and highly conserved protein whose only known function is to serve as a GTPase-Activating Protein (GAP) for Ras. However, most Drosophila NF1 mutant phenotypes, including an overall growth deficiency, are not readily modified by manipulating Ras signaling strength, but are rescued by increasing signaling through the cAMP-dependent protein kinase A pathway. This has led to suggestions that NF1 has distinct Ras- and cAMP-related functions. Here we report that the Drosophila NF1 growth defect reflects a non-cell-autonomous requirement for NF1 in larval neurons that express the R-Ras ortholog Ras2, that NF1 is a GAP for Ras1 and Ras2, and that a functional NF1-GAP catalytic domain is both necessary and sufficient for rescue. Moreover, a Drosophila p120RasGAP ortholog, when expressed in the appropriate cells, can substitute for NF1 in growth regulation. Our results show that loss of NF1 can give rise to non-cell-autonomous developmental defects, implicate aberrant Ras-mediated signaling in larval neurons as the primary cause of the NF1 growth deficiency, and argue against the notion that neurofibromin has separable Ras- and cAMP-related functions.
Keywords: Neurofibromatosis type 1, organismal growth control, non-cell autonomy, Ras signal transduction, Drosophila melanogaster
Neurofibromatosis type 1 (NF1, OMIM 162200) is a common genetic disorder, affecting two to three per 10,000 live births worldwide (Huson and Hughes 1994). NF1 patients are predisposed toward developing a variety of defects, the most characteristic of which include areas of abnormal skin pigmentation and benign tumors associated with peripheral nerves, termed neurofibromas. Less universal but more serious symptoms also include malignant peripheral nerve sheath tumors, other malignancies, and learning disabilities. Developmental abnormalities, such as specific skeletal defects, macrocephaly, and short stature, are also associated with NF1 (Huson and Hughes 1994). The >2800-amino-acid NF1 protein, termed neurofibromin, includes a segment related to the catalytic domains of Ras-specific GTPase-Activating Proteins (GAPs), and ample evidence supports the notion that the ability of neurofibromin to inactivate Ras plays a critical role in the development of NF1-associated tumors (Cichowski and Jacks 2001). The GAP-related domain (GRD) constitutes only ∼15% of neurofibromin, however, and it is less clear whether Ras signaling defects are also the immediate cause of other disease symptoms.
A Drosophila melanogaster NF1 ortholog predicts a protein that is ∼60% identical to human neurofibromin over its entire length. We previously reported that Drosophila NF1-null mutants are viable, fertile, and normally patterned, but display a 15%–20% reduction in linear dimensions during all stages of post-embryonic development (The et al. 1997). NF1 mutants also lack a neuropeptide-stimulated K+ current at the neuromuscular junction (Guo et al. 1997), have a defective escape response (The et al. 1997), display an olfactory learning deficit (Guo et al. 2000), and lack a circadian rest–activity rhythm (Williams et al. 2001). The circadian defect is partially restored by mutations that attenuate Ras signaling (Williams et al. 2001). However, all other analyzed phenotypes lack dosage-sensitive genetic interactions with mutations that alter Ras signaling strength. These Ras-insensitive NF1 phenotypes, however, are suppressed by increasing and enhanced or mimicked by decreasing the activity of the cAMP/PKA signaling pathway (Guo et al. 1997, 2000; The et al. 1997). A functional link between NF1 and cAMP/PKA signaling is further supported by the detection of a reduced cAMP level in Nf1−/− versus Nf1+/− mouse embryos, and by reports of cAMP signaling defects in NF1-deficient fly brain extracts (Tong et al. 2002; Hannan et al. 2006). Arguing that any cAMP/PKA-related function may be evolutionarily conserved, expression of human neurofibromin rescued the Drosophila mutant size defect (Tong et al. 2002).
The only known enzymatic activity of neurofibromin is the ability of its GRD to stimulate the GTPase activity of Ras (Cichowski and Jacks 2001). However, studies in Drosophila and in mammalian cells (Dasgupta et al. 2003) have led to suggestions that neurofibromin may also affect cAMP/PKA signaling, potentially independent of its role as a Ras regulator (Hannan et al. 2006). It is therefore important to determine whether the growth-regulating properties of Drosophila NF1 are separable from its function as a GAP for Ras family GTPases.
In this study, we investigated the cellular and molecular basis of the NF1 size defect. We demonstrate that NF1 function in specific neurons of the larval CNS accounts for its ability to regulate organismal growth. We also demonstrate that this function of NF1 is inseparable from its function as a GAP for Ras family GTPases, arguing against the notion that NF1 has separate Ras- and cAMP-regulating functions. Our studies also implicate the R-Ras ortholog, Ras2, in the pathway by which NF1 regulates growth.
Results
Characterization of new NF1 mutants
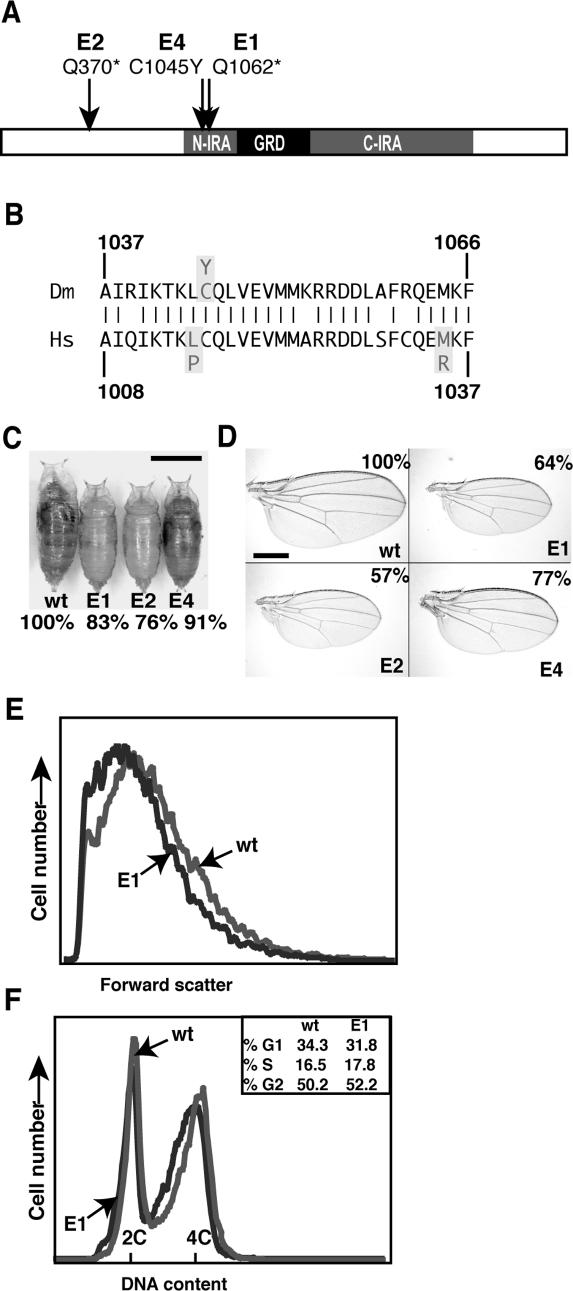
NF1 alleles used in all previous studies were generated by mobilizing a P transposon in a nonisogenic fly strain. Of these original alleles, NF1P1 represented a deletion of most of the NF1 coding region and of at least two genes in the adjacent Enhancer-of-split complex, whereas in NF1P2, a duplicate transposon located in the first NF1 intron interrupted its expression (The et al. 1997). Given that Drosophila NF1 phenotypes are quantitative defects that might be sensitive to genetic background differences, neither allele was ideal for genetic studies. Thus, we used a chromosome 2 and 3 isogenized stock to conduct an F1 screen for ethyl methane sulfonate-induced mutations that failed to complement the NF1 small pupa phenotype. Screening 30,000 pupae yielded three new NF1 alleles. NF1E1 and NF1E2 have nonsense mutations upstream of the catalytic GRD, truncating the protein after 1061 and 369 amino acids, respectively (Fig. 1A). NF1E4 is a C1045Y missense mutation in a conserved part of neurofibromin that also harbors two disease-associated missense mutations (Wu et al. 1996; Kluwe et al. 2003). In sequential immunoprecipitation immunoblot (IP-Western) experiments, using monoclonal antibodies generated against a C-terminal protein segment (The et al. 1997), no NF1 protein was detected in NF1E1 or NF1E2 lysates, whereas NF1E4 and wild-type protein levels were indistinguishable (data not shown).
Figure 1.
Characterization of new NF1 alleles. (A) Location of NF1E1, NF1E2, and NF1E4 mutations. (B) The NF1E4 C1045Y missense mutation occurs in a conserved protein segment that also harbors two disease-associated human missense mutations. (Hs) Homo sapiens; (Dm) D. melanogaster. (C) NF1 pupae are reduced in size. Shown are male pupae of the indicated genotypes. Bar, 1 mm. Male pupal length at 25°C (n > 20)—isogenic wild-type control: 3.01 mm (±0.09); NF1E1: 2.49 mm (±0.07); NF1E2: 2.30 mm (±0.08); NF1E4: 2.53 mm (±0.11); NF1E4 at 18°C: 2.75 mm (±0.11). (D) NF1 wings are reduced in size. Female wings of the indicated genotypes are shown. Bar, 0.5 mm. Wing areas are shown as the percentage of the wild-type control. (E) The mean forward scatter value of dissociated NF1E1 third instar wing disc cells was 86% that of wild-type cells. (F) FACS analysis of propidium-iodide-stained wild-type and NF1E1 third instar wing imaginal disc cells revealed no obvious differences in cell cycle phasing.
Similar to NF1P1 or NF1P2 (The et al. 1997), NF1E1 or NF1E2 pupae are 15%–20% smaller than isogenic wild-type pupae. The NF1E4 missense mutant in this respect behaves as a temperature-sensitive hypomorph (Fig. 1C). Of special relevance to the human disease, several haploinsufficient phenotypes have been described in NF1+/− mammalian cells (Zhu et al. 2002; Wang et al. 2005; Hingtgen et al. 2006). Drosophila NF1 also appears haploinsufficient for growth regulation, since both male and female NF1E1/+ or NF1E2/+ pupae exhibited a small (∼4%), but highly significant (Student t-test p < 0.0001; n = 45) reduction in length compared with isogenic controls (data not shown). The surface area of NF1E1, NF1E2, or NF1E1/NF1E2 adult wings was ∼30%–40% smaller than wings of the parental stock (Fig. 1D). As inferred from the density of wing hairs, this reduction largely reflects a reduction in cell size (data not shown). Wing imaginal discs were similarly reduced in size and made up of smaller cells (Fig. 1E). However, the fraction of wing disc cells in the G1, S, and G2 phases of the cell cycle did not differ appreciably from controls, indicating a proportional reduction in growth during all phases of the cell cycle (Fig. 1F).
NF1 is expressed in post-mitotic larval brain neurons
Previously, NF1−/− epidermal cells generated in the wings of heterozygous animals were found to be of wild-type size, providing the first indication that the requirement for NF1 in regulating growth might be non-cell-autonomous (The et al. 1997). Such nonautonomy could indicate a requirement for NF1 either in cells immediately adjacent to mutant cells, or in more distant cells—possibly even in a different tissue.
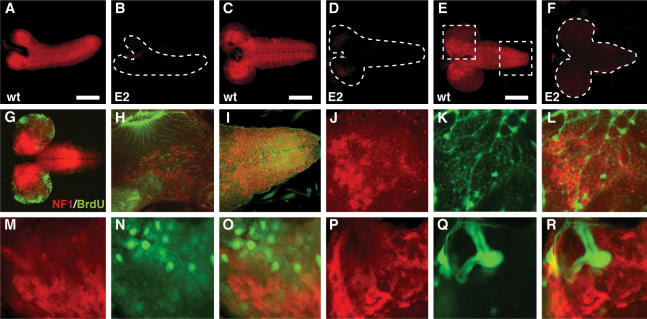
Most insect growth occurs during larval development, and the reduced growth of NF1 mutants first becomes apparent during this phase of the life cycle (The et al. 1997). Immunostaining of dissected wild-type larvae detected little, if any, above background staining in most tissues, including fat body, gut, epidermis or the imaginal discs. The ring gland also lacked obvious staining, which together with other findings argues against a growth-related role for NF1 in this neuroendocrine gland (see below). In contrast, prominent staining was detected in the CNS of wild-type, but not of NF1E1 or NF1E2, larvae. In the CNS of first, second, or third instar larvae, anti-NF1 staining was widespread but not ubiquitous (Fig. 2A–F). In third instar CNS, staining was prominent in the central brain region and in parts of the ventral ganglion, but was low or absent in the proliferative zones of the optic lobes, as witnessed by the lack of overlap between BrdU and NF1 staining (Fig. 2G). Confocal microscopy of third instar CNS revealed complex patterns of intermingled NF1-expressing and -nonexpressing cells (Fig. 2H,I). A lack of overlap between endogenous NF1 expression and UAS-GFP expression driven by the reversed polarity (repo) glial cell GAL4 driver argues that NF1-expressing cells do not represent the glial lineage (Fig. 2J–L). A lack of overlap between endogenous NF1 and grainyhead-driven LacZ expression (Almeida and Bray 2005) similarly argues that NF1-expressing cells are not neuroblasts (Fig. 2M–O). Substantial overlap was observed between endogenous NF1 and elav-GAL4-driven UAS-GFP expression, supporting the notion that NF1-expressing cells are mature neurons (data not shown). Finally, since adult NF1 flies exhibit defective olfactory learning (Guo et al. 2000), and since mushroom bodies (MBs) are neuronal structures implicated in olfactory learning (Skoulakis et al. 1993), it is interesting to note that no obvious NF1 staining was apparent in third instar (Fig. 2P–R) or adult fly brain MBs (data not shown).
Figure 2.
Neurofibromin is expressed in post-mitotic neurons of the larval CNS. (A–F) Confocal images of anti-NF1-stained wild-type (A,C,E) or NF1E2 (B,D,F) first (A,B), second (C,D), or third (E,F) instar CNS. Bars: A,C, 100 μm; E, 200 μm. (G) Lack of overlap between NF1 (red) and BrdU (green) staining. (H,I) Confocal images of the central brain and ventral ganglion regions indicated by hatched boxes in E. (Red) NF1; (green) phalloidin. No obvious differences in staining intensity or in subcellular localization were observed when comparing wild-type and NF1E4 mutants (not shown). (J–L) Lack of overlap between anti-NF1 (red) and repo-GAL4-driven UAS-GFP (green) staining. The region imaged is part of the third instar central brain. (M–O) No overlap between anti-NF1 (red) and grh-lacZ (green) expression in neuroblasts. (P–R) NF1 staining (red) is not obvious in third instar MBs, identified by UAS-GFP expression directed by the 201Y MB GAL4 driver.
NF1 functions in larval neurons to regulate growth non-cell-autonomously
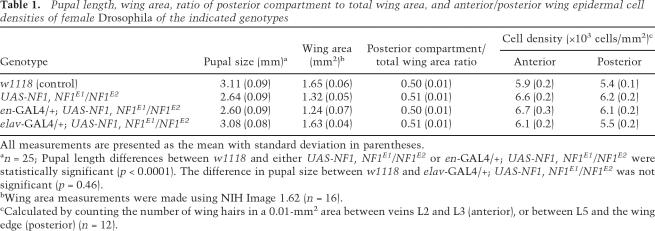
To examine whether NF1 functions in larval neurons to regulate overall organismal growth, we expressed a Drosophila UAS-NF1 transgene in defined larval tissues (Brand and Perrimon 1993). We first analyzed whether expression in wing imaginal discs under the control of the engrailed-GAL4 (en-GAL4) or in larval neurons under the control of the pan-neuronal elav-GAL4 driver was sufficient to rescue the size defect. Staining of third instar imaginal discs and CNS produced the expected patterns of en-GAL4-driven UAS-NF1 expression, restricted to the posterior half of wing discs (Neufeld et al. 1998) and to serotonergic neurons (Lundell et al. 1996) in the CNS (Supplementary Fig. 1). This expression pattern was insufficient to rescue NF1E1/E2 pupal or adult wing size defects (Table 1). Moreover, there was no difference in the relative size of anterior and posterior adult wing compartments, indicating that NF1 expression in a significant portion of the wing disc was unable to affect disc growth. In contrast, elav-GAL4-driven neuronal expression of UAS-NF1 strongly rescued the reduced pupal size phenotype as well as adult wing size defects (Table 1).
Table 1.
Pupal length, wing area, ratio of posterior compartment to total wing area, and anterior/posterior wing epidermal cell densities of female Drosophila of the indicated genotypes
All measurements are presented as the mean with standard deviation in parentheses.
an = 25; Pupal length differences between w1118 and either UAS-NF1, NF1E1/NF1E2 or en-GAL4/+; UAS-NF1, NF1E1/NF1E2 were statistically significant (p < 0.0001). The difference in pupal size between w1118 and elav-GAL4/+; UAS-NF1, NF1E1/NF1E2 was not significant (p = 0.46).
bWing area measurements were made using NIH Image 1.62 (n = 16).
cCalculated by counting the number of wing hairs in a 0.01-mm2 area between veins L2 and L3 (anterior), or between L5 and the wing edge (posterior) (n = 12).
As expected, elav-GAL4-driven UAS-NF1 expression was widespread throughout the larval CNS, but low-level staining was also apparent in the part of the wing disc that gives rise to the wing hinge (Supplementary Fig. 1). Thus, to examine in more detail which tissues or cells require NF1 to support normal growth, we analyzed the rescuing ability of >100 additional GAL4 drivers. For many drivers, we analyzed overall pupal size and adult wing size in parallel with their larval expression pattern, the latter either by staining UAS-NF1 transgenics with NF1 antibodies or by using a UAS-GFP reporter. Drivers that express in the fat body, salivary glands, gut, imaginal discs, lymph gland, or epidermis did not rescue the growth defects (Supplementary Table 1). The common denominator among 16 drivers that did significantly rescue was different degrees of expression in the larval CNS (Supplementary Table 1). Expression of UAS-NF1 directed by repo-GAL4 did not modify NF1 size, arguing against a role for glial cells. Several subsets of neurons can similarly be ruled out as uniquely responsible. It was shown recently that increased Ras signaling in the ecdysone-producing prothoracic gland, which is part of the neuroendocrine ring gland, reduces the overall size of Drosophila (Caldwell et al. 2005; Colombani et al. 2005; Mirth et al. 2005). However, UAS-NF1 expression controlled by the Phantom-GAL4, Aug 21, or P0206-GAL4 ring gland drivers did not rescue the NF1 size defect (Supplementary Table 1). Similarly, Feb 211-, Mai 301-, or Mai 369-driven UAS-NF1 expression in subsets of neurons that innervate the ring gland (Siegmund and Korge 2001) did not restore normal growth. Among the drivers expressed in peptidergic neurons, only the relatively widely expressed GAL4-386Y driver (Taghert et al. 2001) allowed partial rescue. No rescue was observed upon expressing UAS-NF1 in dopaminergic neurons using two Ddc-GAL4 drivers (Li et al. 2000), in cholinergic neurons using Cha-GAL4 19B (Salvaterra and Kitamoto 2001), in amnesiac-expressing cells using the amnc651 driver (Waddell et al. 2000), or in insulin-producing neurosecretory cells using dILP2-GAL4 (Rulifson et al. 2002).
In summary, our experiments demonstrate a role for NF1 in the larval brain to regulate the growth of larval tissues, including wing imaginal discs. Moreover, since NF1 expression in neuronal subpopulations previously implicated in nonautonomous growth control does not restore mutant growth, our findings imply a role for other portions of the larval brain in regulating organismal growth. We have further localized this function to cells in the brain that express the Ras family GTPase Ras2 (see below).
Loss of NF1 enhances CNS MEK/ERK activity, without causing obvious changes in proliferation or differentiation
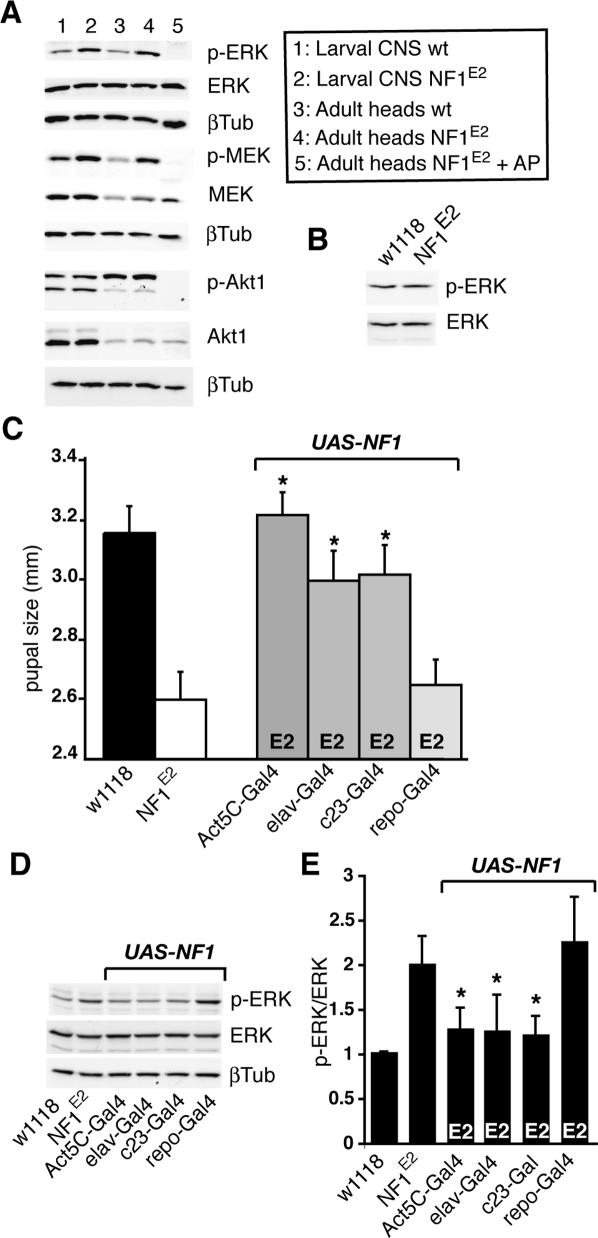
Since the only established biochemical function of neurofibromin is its ability to act as a GAP for Ras (Cichowski and Jacks 2001), we analyzed NF1-deficient third instar larval CNS and adult fly heads for Ras signaling defects. Extending a previous finding (Williams et al. 2001), and consistent with a role as a negative regulator of the Ras–Raf–MEK–ERK cascade, we detected a reproducible two- to fourfold increase in the level of phosphorylated rl ERK kinase (hereafter referred to as p-ERK) in NF1 third instar larval CNS extracts (Fig. 3A, lanes 1,2). Elevated p-ERK was also apparent in adult NF1 fly heads (Fig. 3A, lanes 3,4) but not in wing discs (Fig. 3B). The kinase acting upstream of ERK, Dsor1, showed a similar increase in phosphorylation (Fig. 3A). In contrast, using an assay that detected elevated phospho-Akt1 (p-Akt1) in flies expressing activated Ras1 (Colombani et al. 2005), we observed no change in p-Akt1 levels between NF1 and wild-type larval or adult CNS (Fig. 3A). Arguing that loss of NF1 causes no major defects in cell proliferation or differentiation, confocal analysis of BrdU-stained wild-type and mutant larval CNS revealed no obvious differences in the number or localization of proliferating cells, and expression of several differentiation markers also appeared unchanged (data not shown). Interestingly, anti-NF1 and anti-p-ERK staining overlapped extensively in wild-type CNS (Supplementary Fig. 2A–F). Loss of NF1 increased the intensity of p-ERK staining, but not its pattern (data not shown).
Figure 3.
Enhanced signaling through the MEK/ERK pathway is rescued by expressing UAS-NF1 in neurons. (A) Western blot showing elevated p-ERK and p-MEK, but not p-Akt1, in NF1-deficient third instar or adult CNS compared with w1118 controls. Lysates were prepared from w1118 or NF1E2 larval CNS (lanes 1,2), from w1118 (lane 3), or from NF1E2 (lanes 4,5) adult heads. The extract in lane 5 was incubated for 15 min at 37°C with 20 U of alkaline phosphatase (Roche Diagnostics) prior to electrophoresis. (B) p-ERK levels are not elevated in NF1-deficient wing discs. (C) Rescue of female pupal size (n = 30) upon expression of UAS-NF1 under the control of the ubiquitous Act5C-GAL4 or the neuronal elav-GAL4 or c23-GAL4 drivers, but not upon repo-GAL4-driven glial cell expression rescue. (*) p < 0.0001, Student t-test. (D) Pupal size rescue correlates with rescue of the third instar CNS p-ERK phenotype. (E) Third instar CNS p-ERK/ERK ratio determined by densitometric scanning of films from three experiments. (*) p < 0.05, Student t-test.
Neuronal expression of a functional NF1GRD is necessary and sufficient for size rescue
Ubiquitous or neuronal UAS-NF1 expression rescued NF1 pupal size, whereas glial expression did not (Fig. 3C). Size rescue correlates with suppression of the elevated third instar CNS p-ERK phenotype (Fig. 3D,E). Taken together, these findings are consistent with NF1 functioning as a GAP for a Ras family GTPase in specific parts of the brain to regulate growth. However, it has been suggested that the NF1 protein may have functions independent of its RasGAP activity, and these may be required for its ability to regulate organismal growth (Hannan et al. 2006). To distinguish between these possibilities, we tested whether the catalytic GRD was required for size rescue, and whether other protein segments were also essential.
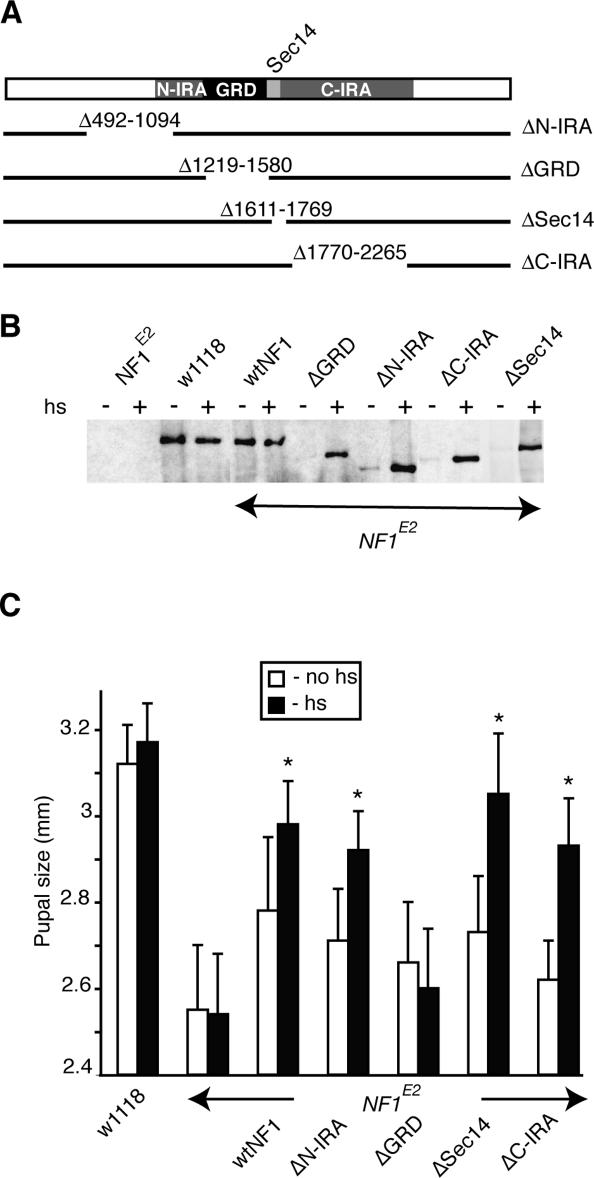
Neurofibromin shares ∼20% sequence identity with the budding yeast Ira1p and Ira2p RasGAPs over approximately half its length. The Ira-related segment includes the GRD and a flanking Sec14-pleckstrin homology putative lipid-binding domain (Aravind et al. 1999; D’Angelo et al. 2006). To determine which parts of neurofibromin are essential for rescuing the size defect, we generated heat-shock-inducible Drosophila NF1 transgenes bearing in-frame deletions (Fig. 4A). Flies expressing these transgenes were crossed into the NF1E2 background and at least two transgenic lines expressing similar protein levels were analyzed for each construct (Fig. 4B).
Figure 4.
Large segments of neurofibromin other than the GRD are dispensable for size rescue. (A) Extent of four in-frame deletions. (B) IP-Western analysis of adult head extracts showing different levels of heat-shock-inducible (hs) transgene expression. (C) Female pupal size of flies of the indicated genotypes, which did (filled-in columns) or did not (open columns) receive daily heat shocks. The graph shows the average female pupal length and the standard deviation calculated by measuring >20 pupae for each genotype. Asterisks indicate significant rescue (Student t-test, p < 0.0001).
We previously described two Drosophila NF1 splice forms with different C termini and reported that expression of the shorter protein restored NF1 mutant growth (The et al. 1997). Subsequent analysis revealed that the rescuing transgene also lacked alternatively spliced exon 14, coding for amino acids 2548–2577. Interestingly, the position of Drosophila NF1 exon 14 corresponds almost exactly to where exon 43 is alternatively spliced in human NF1 (Vandenbroucke et al. 2002). However, a hsp70-NF12802 transgene that included exon 14 and the longer C-terminal exon rescued pupal size to the same extent as the original hsp70-NF12734 transgene (data not shown). Thus, alternative splicing of C-terminal exons does not affect size rescue.
Surprisingly, deletions that remove large parts of neurofibromin other than the GRD were able to rescue the size defect (Fig. 4C). These included a number of protein segments that are highly conserved between Drosophila and human NF1. The Sec14 domain, which was recently suggested to harbor three potential caveolin-binding sites (Boyanapalli et al. 2006), is entirely dispensable for size rescue. The Δ1770–2265 mutant, which lacks most of the remainder of the C-terminal Ira-related segment, including a recently described pleckstrin-like domain, also rescued the size defect—albeit that efficient rescue required transgene homozygosity. A large proportion of disease-associated NF1 missense mutations occur in a region upstream of the Ira-related segment, suggesting the existence of a second functional domain (Fahsold et al. 2000; Mattocks et al. 2004). However, the Δ492–1092 mutant, engineered to remove the region corresponding to this upstream mutation cluster, also rescued. In contrast, the Δ1219–1580 GRD deletion mutant was expressed, but did not rescue growth (Fig. 4).
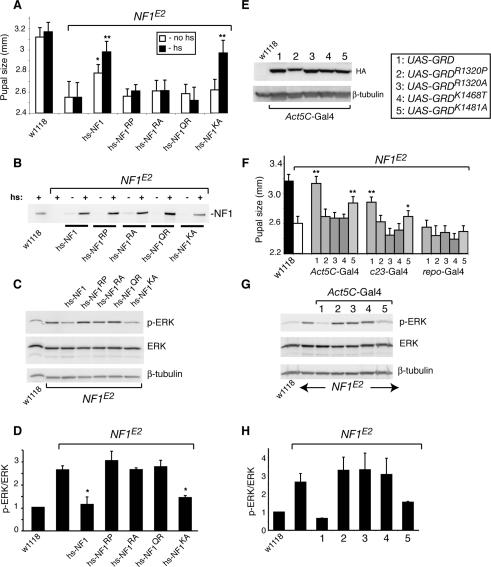
Since the GRD appears necessary for rescue, we next analyzed whether GAP activity was required. To this end we generated flies expressing four transgenes harboring single amino acid substitutions predicted to interfere with GAP activity. In two mutants, the catalytically essential Arg 1320 in the GRD finger loop (Scheffzek et al. 1997) was substituted for either a proline or an alanine. The corresponding human R1276P and R1276A mutants have >1000-fold reduced GAP activity (Klose et al. 1998; Sermon et al. 1998). In a third mutant, Gln 1471 in the α7/variable loop of the GRD was substituted for an arginine. The corresponding Q1426R human mutant behaves as a loss-of-function mutant in a yeast Ira complementation assay (Gutmann et al. 1993). Finally, to address the concern that catalytically impaired mutants might attenuate Ras signaling by sequestering the GTPase, we also generated a K1481A mutant. Lys 1481 does not map near the catalytic site, but undergoes electrostatic interactions with charged residues in the switch 1 region of Ras (Scheffzek et al. 1997). The corresponding K1436A human neurofibromin mutant had 96-fold reduced affinity for Ras, but near normal catalytic activity at saturation (Ahmadian et al. 2003). All mutant proteins were expressed at similar levels (Fig. 5B), but only the catalytically active K1481A mutant rescued pupal size and third instar CNS p-ERK levels (Fig. 5A,C,D).
Figure 5.
Expression of a functional NF1GRD is necessary and sufficient for rescuing pupal size and larval CNS p-ERK phenotypes. (A) Average female pupal size with (filled columns) or without (open columns) heat-shock induction of the indicated transgenes. Asterisks indicate significant rescue. (*) P < 0.001; (**) P < 0.0001; Student t-test. (B) IP-Western analysis showing similar levels of heat-shock-inducible transgene expression. (C) Rescue of NF1 pupal length correlates with decreased p-ERK levels in third instar larval CNS. (D) Graph showing average p-ERK/ERK ratios of three experiments. (*) p < 0.025, Student t-test. (E) Western blot showing similar levels of Act5C-GAL4-driven wild-type and mutant UAS-GRD expression in adult flies. The R1230P mutant includes 10 extra amino acids following its HA tag, explaining its larger size. The boxed legend indicates the transgenes used in lanes/columns 1–5 in E–H. (F) Ubiquitous or neuronal, but not glial, expression of wild-type UAS-GRD suffices to rescue pupal size. Of four GAP-impaired mutants, only K1481A gave rise to partial rescue, similar to results obtained with full-length transgenes. (**) p < 0.0001; (*) p < 0.001; Student t-test. (G) Pupal size rescue correlates with the ability of UAS-GRD transgenes to reduce adult head p-ERK levels. (H) Average third instar CNS p-ERK/ERK ratios observed in two experiments.
Since GAP activity appears essential for rescue, we next asked whether expression of a truncated NF1GRD protein was sufficient. We also tested four NF1GRD constructs bearing R1320A, R1320P, K1468T, and K1481A mutations. K1468T was the only mutant not tested in the context of a full-length transgene. K1468 corresponds to human NF1 K1423, implicated as important for GAP activity and microtubule association of neurofibromin (Poullet et al. 1994; Xu and Gutmann 1997). All proteins were expressed at similar levels when driven by Act5C-GAL4 (Fig. 5E). Demonstrating that the NF1GRD is sufficient for rescue, the wild-type protein strongly rescued pupal size. Significant rescue was also observed when the wild-type protein was expressed in neurons using c23-GAL4, but not upon expression in glial cells using repo-GAL4 (Fig. 5F). Three of four GRD missense mutants failed to rescue size or p-ERK phenotypes, either when expressed in neurons or ubiquitously. Similar to the full-length K1481A transgene, the K1481A NF1GRD mutant rescued both phenotypes partially (Fig. 5F–H). Thus, expression of a functional NF1-GAP catalytic domain is both necessary and sufficient to restore normal growth.
NF1 expression in Ras2-expressing cells rescues size and ERK activation defects
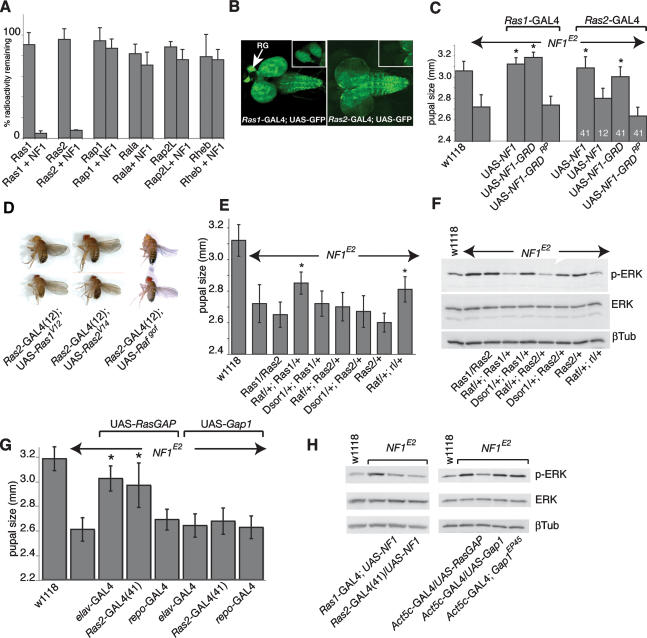
Expression of a functional NF1GRD is necessary and sufficient for size rescue, yet in previous studies several Ras pathway mutants did not dominantly modify NF1 size (The et al. 1997; Williams et al. 2001). This raised the possibility that NF1 could also act as a GAP for other Ras-like GTPases and that this role for NF1 might be important in its ability to regulate growth. Human NF1 is a GAP for H-Ras, K-Ras, and N-Ras, and for all three R-Ras paralogs (Rey et al. 1994; Ohba et al. 2000; Huang et al. 2004). In Drosophila, Ras1 is orthologous to H-Ras, K-Ras, and N-Ras, whereas Ras2 is most similar to mammalian R-Ras paralogs. In biochemical GAP activity assays, a bacterially produced Drosophila NF1GRD protein strongly enhanced GTP hydrolysis by Drosophila Ras1 and Ras2, but not by Rap1, Rap2L, Rala, or Rheb (Fig. 6A).
Figure 6.
Defective Ras regulation in Ras2-expressing neurons underlies the size defect. (A) Drosophila NF1 is a GAP for Ras1 and Ras2, but not for Rap1, Rap2L, Rala, or Rheb. The graph shows the percentage GTPase-bound γ32P-GTP remaining after a 10-min incubation with purified NF1GRD protein. (B) Third instar CNS Ras1-GAL4 and Ras2-GAL4(41)-driven UAS-GFP expression. The inserts show higher-magnification views of the ring glands. (C) Wild-type but not GAP-deficient UAS-NF1GRD expression driven by Ras1-GAL4 or by the strong Ras2-GAL4(41) driver rescues female pupal size. (*) p < 0.0001, Mann-Whitney test. (D) Ras2-GAL4(12)-driven UAS-Ras1Val12, UAS-Ras2Val14, or UAS-Rafgof expression phenocopies NF1 size when grown at 16°C. The flies on top lack the Ras2-GAL4 driver. One strong and one weak UAS-Ras1Val12 line were tested. Both reduced size, but only the weaker line resulted in viable flies. (E) Partial rescue of NF1E2 female pupal size by combined heterozygous loss of Ras1 and Raf, Ras2 and Raf, or Raf and rl. (*) p < 0.0001, Mann-Whitney. (F) Three double-mutant combinations fully restore elevated larval p-ERK levels. (G) Rescue of NF1E2 female pupal size defect by elav-GAL4- or Ras2-GAL4-driven UAS-RasGAP but not UAS-Gap1 expression. (*) p < 0.0001, Mann-Whitney. (H) Expression of UAS-NF1 and UAS-RasGAP but not UAS-Gap1 or Gap1EP45 rescues the elevated CNS p-ERK phenotype.
A failure to isolate mutants in a large F2 lethal screen previously suggested that loss of Drosophila Ras2 might not be lethal (Harrison et al. 1995). Ras2 is expressed from a promoter that also controls the expression of the SEC1-related Ras opposite, or Rop, gene (Salzberg et al. 1993). Ras2/Rop promoter-driven LacZ expression was previously detected in third instar CNS (Salzberg et al. 1993), in a pattern resembling the NF1 expression pattern. Thus, to determine whether expression of UAS-NF1 in Ras2-expressing cells suffices to rescue, we generated transgenic lines expressing GAL4 under the control of the Ras2/Rop promoter. Among two Ras2-GAL4 lines used, Ras2-GAL4(41) gives rise to high levels of UAS-GFP expression in salivary glands, in the gut, and in a specific pattern within the central brain region and the ventral ganglion of the larval CNS, consistent with previous results (Salzberg et al. 1993). The Ras2-GAL4(12) line drove expression in essentially the same pattern, but at a lower level. The third instar CNS Ras2-GAL4(12)-driven UAS-GFP expression pattern is shown in Figure 6B. Confocal analysis revealed substantial but incomplete overlap between Ras2-GAL4-driven UAS-GFP and endogenous NF1 expression (Supplementary Fig. 2). We also generated Ras1-GAL4 transgenics. As expected, Ras1-GAL4 drivers caused widespread UAS-GFP expression throughout development, but in third instar CNS, expression appeared especially high in the ring gland, whereas the Ras2 driver was not detectably expressed in this tissue (Fig. 6B). Expression of UAS-NF1 or UAS-NF1GRD transgenes directed by the strong but not the weak Ras2-GAL4 driver potently rescued the size defect, suggesting that that the NF1 size defect reflects a function for neurofibromin in Ras2-expressing cells. Not unexpectedly, rescue was also observed when UAS-NF1 was driven with the widespread Ras1-GAL4 driver (Fig. 6C). Compatible with the notion that elevated Ras activity in Ras2-expressing cells contributes to the NF1 size defect, expression of either UAS-Ras1Val12 or UAS-Ras2Val14 under the control of the weak Ras2-GAL4(12) driver resulted in small dead pupae when cultures were maintained at 25°C or 18°C, whereas flies that eclosed at 16°C phenocopied the NF1 size defect. Expression of a gain-of-function Raf mutant in Ras2-expressing cells similarly phenocopied the NF1 size defect (Fig. 6D; Supplementary Table 2). Taken together, these results imply that the level of Ras pathway signaling in Ras2-expressing cells is important in regulating organismal size.
Dosage-sensitive genetic suppression of NF1 phenotypes by Ras pathway mutants
Previously, we showed that two Ras1 loss-of-function alleles did not dominantly modify NF1 size phenotypes (The et al. 1997). It is possible, however, that heterozygous loss of Ras1 is insufficient to restore normal signaling in an NF1-null mutant background. Thus, we analyzed modification of pupal size and larval p-ERK phenotypes using multiple single- and double-mutant combinations in both NF1E2-null and NF1E4 hypomorphic mutants. Heterozygous loss of Ras1 or of Ras2 [using Df(3L)GN34 and Df(3L)GN19, which uncover Ras2], or combined loss of Ras1 and Ras2 was insufficient to modify pupal size or larval p-ERK phenotypes of either NF1E2 (Fig. 6E,F) or NF1E4 (data not shown). All tested single mutants affecting canonical Ras effectors similarly did not modify NF1 pupal size. Tested mutants include alleles of RalGDS ortholog Rgl, its target GTPase Rala, and PI3K21B, encoding the p60 regulatory subunit of Drosophila class 1 PI3 kinase. No modification of NF1 size was observed upon Ras2-GAL4(41)-driven expression of a UAS-Dp110D954A dominant-negative PI3 kinase transgene (Leevers et al. 1996), and Ras2-GAL4(41)-driven expression of a constitutively active UAS-Dp110CAAX transgene did not phenocopy NF1 size (data not shown). Similarly, Ras2-GAL4(41)-driven expression of either dominant-negative Ras1 or dominant-negative Raf did not rescue NF1 size phenotypes. The mTor pathway is activated in NF1-deficient mammalian cells (Dasgupta et al. 2005; Johannessen et al. 2005), but two Tor alleles did not modify size. Loss-of-function mutants affecting raf, Dsor1, and rl—components of the canonical Raf–MEK–ERK cascade—also did not modify (Supplementary Fig. 3). However, combined heterozygous loss of raf and rl, Ras1 and raf, or Ras2 and raf fully rescued the larval CNS p-ERK phenotype (Fig. 6F), while the former two double-mutant combinations also rescued pupal size, but only partially (Fig. 6E). These observations indicate that reducing Ras pathway signaling to achieve a wild-type level of ERK activation is in itself insufficient to rescue the pupal size phenotype. This suggests that other effector mechanisms may be even more sensitive to the levels or to the kinetics of Ras pathway activation.
If unregulated signaling through one or more Ras1 and/or Ras2 effectors explains the NF1 size defect, other Drosophila RasGAPs that differ considerably from NF1 outside the GRD could potentially substitute for NF1. Thus, we analyzed whether expression of p120RasGAP ortholog RasGAP (Feldmann et al. 1999) or Gap1 (Gaul et al. 1992) modified NF1 size. As shown in Figure 6, G and H, elav-GAL4 or Ras2-GAL4(41), but not repo-GAL4-driven UAS-RasGAP expression suppressed the NF1 size defect, whereas Act5C-GAL4-driven expression also suppressed the elevated larval CNS p-ERK level. Interestingly, expression of Gap1EP45 (Rorth 1996) did not rescue either defect, even though GMR-GAL4-driven Gap1EP45 expression gave rise to the previously characterized (Rorth 1996) Ras1-dependent rough eye defect (data not shown). The inability of Gap1EP45 to rescue does not appear to reflect insufficient expression, since identical results were obtained with transgenic lines expressing a GAL4-inducible Gap1 transgene, specifically generated for this purpose. Similar to Gap1EP45, eye-specific expression of UAS-Gap1 caused rough eye phenotypes, whereas ubiquitous expression was lethal. Neuronal expression driven by elav-GAL4 or Ras2-Gal4(41) was not lethal, but did not rescue NF1 size or p-ERK defects (Fig. 6G,H). Thus, rescue of size and p-ERK phenotypes appears to reflect a property shared between Drosophila NF1 and RasGAP, but not Gap1.
Discussion
Enhancing the GTPase activity of Ras family members is the only known biochemical activity of neurofibromin, the protein defective in patients with NF1 (Cichowski and Jacks 2001). This has focused much attention on manipulating Ras signaling as a way to correct the diverse symptoms of NF1. However, most Drosophila NF1 phenotypes lack dosage-sensitive genetic interactions with mutants that affect signaling by Ras1, the single fly ortholog of mammalian H-Ras, K-Ras, and N-Ras. Rather, an NF1 mutant growth deficiency, an electrophysiological defect, and a defect in olfactory learning are rescued by manipulations that increase signaling through the cAMP/PKA pathway (Guo et al. 1997, 2000; The et al. 1997). These findings have led to suggestions that neurofibromin may affect cAMP/PKA signaling in a Ras-independent manner, a hypothesis supported by a recent report that human NF1 suppresses Drosophila NF1 mutant size independent of GAP activity (Hannan et al. 2006). In contrast, our experiments using Drosophila NF1 transgenes suggest that loss of RasGAP activity is inseparable from the NF1 size defect. The reason for this discrepancy remains unclear, but may reflect inappropriate interactions between human neurofibromin and Drosophila GTPases or other proteins involved in growth regulation.
Our results show that the impaired growth of Drosophila mutants reflects a non-cell-autonomous role for NF1 in larval neurons. While runting is relatively common in mutant mice, we note that mice engineered to specifically lack neuronal Nf1 expression were previously also found to be small (Zhu et al. 2001). Growth in Drosophila proceeds during three larval instars that culminate in pupariation, pupation, and adult eclosion. As in other animals, growth is affected by feeding, which in Drosophila occurs during the first two and most of the third larval instar. Early in the third instar, larvae reach what is known as critical weight, a point at which holometabolous insects commit to metamorphosis and can develop without further feeding (Beadle et al. 1938; Davidowitz et al. 2003). Two neuroendocrine pathways have been implicated in coordinating feeding with Drosophila development and overall growth, but our results argue against obvious roles for NF1 in either one. Perhaps the best-understood growth-related pathway involves Drosophila insulin-like proteins (dILPs), three of which are produced—two in a nutrient-dependent manner—by bilateral symmetric groups of seven neurosecretory cells in the pars intercerebralis of the larval CNS (Ikeya et al. 2002). Ablating these cells causes a severe growth defect that is rescued by expression of a dILP2 transgene (Rulifson et al. 2002). In peripheral tissues, dILPs activate the insulin receptor, leading to the phosphorylation of CHICO and the recruitment of a class I PI3 kinase, consisting of Dp110 catalytic and p60 regulatory subunits. Genetic manipulations that increase signaling through this pathway increase the size of peripheral tissues in a cell-autonomous manner, whereas loss-of-function mutations have the opposite effect (Chen et al. 1996; Bohni et al. 1999; Weinkove et al. 1999). Recently, insulin was found to control developmental timing, but not body or organ size, during the period before Drosophila achieves critical weight, whereas after reaching this set point insulin no longer affected developmental timing, but only body and organ size (Shingleton et al. 2005). Our analysis of mutant development and behavior, which will be reported elsewhere, found no differences in feeding or developmental timing between NF1 mutants and isogenic controls. Moreover, the lack of dosage-sensitive genetic interactions between NF1 and PI3 kinase p60 or Tor mutants, and the observation that dILP2-GAL4-driven UAS-NF1 expression in insulin-producing neuroendocrine cells does not modify NF1 size, all argue that insulin deficiency is not likely to be a major contributor to the NF1 size defect.
Drosophila growth and development are also coordinated by a hormonal cascade involving juvenile hormone (JH), prothoracicotrophic hormone (PTTH), and ecdysone. JH and ecdysone are produced by the corpora allata and the thoracic gland, respectively, which together with the corpora cardiaca form the neuroendocrine ring gland. PTTH stimulates ecdysone release and is made by neurons that innervate the thoracic gland in response to a developmentally controlled reduction in JH titer. JH production, in turn, is controlled by insulin, explaining the developmental delay and increased longevity of some hypomorphic insulin pathway mutants (Tatar et al. 2001). Three groups recently reported that increasing the size of the prothoracic gland by manipulations that activate Ras1 or its Dp110 PI3 kinase effector impairs Drosophila growth (Caldwell et al. 2005; Colombani et al. 2005; Mirth et al. 2005), possibly through ecdysone-mediated attenuation of insulin signaling in peripheral tissues (Colombani et al. 2005). Again, our inability to modify NF1 size by expressing UAS-NF1 in the prothoracic gland, in other parts of the ring gland, or in neurons that innervate the ring gland suggests that excess Ras activity resulting from a loss of NF1 in these cells or tissues does not provide an easy explanation for the impaired growth of NF1 mutants. Further arguing against such a role, no obvious NF1 expression was detected in the ring gland.
Ras2-GAL4 is among the most restricted drivers that rescue NF1 size when driving UAS-NF1. This fact, combined with the observation that neuronal but not glial drivers similarly rescue, suggests that Ras2-GAL4-expressing cells are neuronal. It remains unclear in what proportion of these cells NF1 is required to restore growth, but costaining experiments revealed substantial overlap between endogenous NF1 and Ras2-GAL4-driven UAS-GFP expression. Moreover, Ras2-GAL4-driven UAS-NF1 expression strongly suppressed the larval CNS p-ERK phenotype. Several other findings support our conclusion that a Ras signaling defect in Ras2-GAL4-expressing cells is the primary cause of the NF1 size defect. First, Ras2-GAL4-driven expression of a functional NF1GRD is necessary and sufficient for rescue. Second, Ras2-GAL4-driven expression of activated Ras1 or Ras2 phenocopied the NF1 size defect. Third, Ras2-GAL4-driven expression of a Drosophila p120RasGAP ortholog also rescued, arguing that the ability to rescue reflects a property shared between NF1 and RasGAP. Interestingly, expression of a third Drosophila RasGAP, Gap1, did not rescue either size or p-ERK phenotypes. Whether the inability of Gap1 to substitute for NF1 reflects an inappropriate expression level or some other factor—such as different regulation, localization, or GTPase substrate specificity—remains to be determined.
Initial reports that increasing cAMP/PKA activity rescued Drosophila NF1 phenotypes generated much interest, in part because cAMP plays a prominent role in learning, which is impaired in many children with NF1. However, subsequent studies showed that genetic or pharmacologic manipulations that attenuate Ras signaling restored learning in heterozygous Nf1 mutant mice (Costa et al. 2002). Altered Ras signaling in the CNS appears capable of regulating the growth of the larval epidermis and imaginal discs. This could occur by modulating the levels of diffusible growth factors or growth inhibitors. Conceivably, cAMP/PKA signaling could be of importance at a more downstream component of this pathway, such as the release of, or response to, such diffusible factors.
Our results also demonstrate that heterozygous loss of individual genes encoding canonical Ras pathway components is insufficient to restore p-ERK activity in homozygous null or hypomorphic NF1 mutants. Interestingly, combined loss of Raf and rl, Ras1 and Raf, and Ras2 and Raf fully rescued the larval p-ERK defect, while the former two double mutants partially restored pupal size. Thus, Ras1 and Ras2 may jointly contribute to ERK activation in NF1-deficient CNS. Whether Ras effectors other than Raf/ERK contribute to the NF1 size defect, and how enhanced PKA activity rescues NF1 phenotypes remain to be determined.
Materials and methods
Fly stocks
Flies were maintained on standard agar–oatmeal–molasses medium at 25°C, unless otherwise specified. The following mutant and transgenic fly strains were used: P[hsp70-NF12734] (The et al. 1997), P[hsp70-NF1Δ492–1094], P[hsp70-NF1Δ1219–1580], P[hsp70-NF1Δ1611–1769], P[hsp70-NF1Δ1770–2265], P[hsp70-NF1R1320A], P[hsp70-NF1R1320P], P[hsp70-NF1Q1471R], P[hsp70-NF1K1481A], P[hsp70-NF12802], P[UAS-NF1], P[UAS-NF1GRD], P[UAS-NF1GRDR1320A], P[UAS-NF1GRDR1320P], P[UAS-NF1GRDK1468T], P[UAS-NF1GRDK1481A], P[Ras1-GAL4], P[Ras2-GAL4], P[UAS-Gap1] (this study), P[UAS-GFP] (Yeh et al. 1995), P[UAS-Ras1V12] (M. Go and S. Artavanis-Tsakonas, unpubl.), P[UAS-Ras2V14] (Brand and Perrimon 1993), P[UAS-RasGAP] (Feldmann et al. 1999), Gap1EP45 (Rorth 1996), P[UAS-Dp110D954A], P[UAS-Dp110CAAX] (Leevers et al. 1996), Ras1e1B, Ras1e2F, RglBG02025, RalaG0174, RalaKG06114, PI3K21BEY06407, P[UAS-Raf-gof]F179, P[lacW]grhS2140, TorK17004 and TorΔP, phl12, Dsor1S1221, and rl1. Mutants for which no references are provided were obtained from the Bloomington stock center. Supplementary Table 1 gives the origin of GAL4 driver lines.
NF1 mutagenesis screen
NF1 alleles were generated by crossing ethyl methane sulfonate mutagenized second and third chromosome isogenized w1118 males to NF1P2 females. Screening 30,000 F1 pupae identified three dominant mutations resembling the Tubby mutant, and four recessive potential new NF1 alleles. NF1E3 represented a deletion and was discarded. The NF1E1, NF1E2, and NF1E4 coding sequences were PCR-amplified and sequenced. Any detected mutation was verified by analyzing independent PCR products.
Transgenic rescue
Transgenic rescue experiments were performed using cultures maintained on freshly prepared food at similar density. For pupal size measurements, >50 pupae of each genotype were measured using a video-equipped microscope. Pupae were then allowed to eclose, and measurements for >20 male or female pupae were used to calculate average size, standard deviations, and statistical significance. To allow for slight variations, controls were included in each experiment. Wing surface areas were determined using NIH Image 1.62 software. Wing cell density was determined by counting the number of hairs in 0.01-mm2 areas between the L2 and L3 veins, and between the L5 vein and the wing edge. Hsp70 promoter-containing transgenes were induced by a daily 30 min heat shock at 37°C. Wing disc cell size was determined using a Cytomation MoFlo cytometer (Neufeld et al. 1998), and data were analyzed using FloJo software (Tree Star, Inc).
Transgenes
UAS-NF1 was made by transferring the insert of a hsp70-NF1 mini-gene (The et al. 1997) into the pUAS-T vector (Brand and Perrimon 1993). This transgene includes the shorter C terminus predicted by exon 18b (The et al. 1997) and lacks a 30-amino-acid segment predicted by alternatively spliced exon 14. A hsp70-NF12802 transgene that includes exon 14 and the longer C-terminal segment predicted by exon 18a was generated by standard cloning. Missense and deletion mutants were generated by PCR-based mutagenesis. UAS-NF1GRD transgenes were engineered to include an AUG codon upstream of amino acids 1214–1574, followed by a HA tag and a termination codon. Ras1-GAL4 and Ras2-GAL4 drivers were generated by directionally cloning PCR-amplified 1850 (Ras1) and 358-base-pair (Ras2) genomic segments representing the presumed transcriptional promoters into the pChs-Gal4 vector (a gift from Dr. Holger Apitz). The insert for the UAS-Gap1 transgene was generated by PCR amplification of first strand cDNA and cloned into pUAS-T (Brand and Perrimon 1993). All constructs were sequenced prior to embryo injection. Transgenic flies were generated by standard procedures. Further details about constructs are available upon request.
GAP activity assay
A plasmid encoding a maltose-binding protein NF1GRD fusion protein was made by cloning a PCR-amplified Drosophila NF1 cDNA segment encoding amino acids 1236–1594 into the pMal-c2X (New England Biolabs) vector. cDNAs for Ras1 (CG9375), Ras2 (CG1167), Rap1 (CG1956), Rap2L (CG3204), Rala (CG2849), and Rheb (CG1081) were PCR-amplified from first strand cDNA and similarly cloned into pMal-c2X. All insert sequences were verified. Soluble fusion proteins were affinity-purified on Amylose resin. Active GTPase concentrations were determined by α32P-GTP binding and GAP activity assays performed as described, using 6 nM active GTPase per reaction (Brill et al. 1996).
Miscellaneous techniques
Larvae were dissected in PBS, fixed in 4% paraformaldehyde, and permeabilized using 0.1% Triton X-100 in PBS. Larval tissues were stained with monoclonal antibody DNF1-21 (The et al. 1997) and anti-mouse-Cy5 secondary antibody and viewed using a Zeiss LM510 confocal microscope. For Western blot analysis, larval CNS was dissected, collected on dry ice, and homogenized in lysis buffer (100 mM NaCl, 10 mM Tris at pH 7.6, 1 mM EDTA, 1% Triton X-100, 10 mM β-glycerolphosphate, 10 mM NaF; 1 mM Na3VO4). Adult fly heads were prepared as described (Williams et al. 2001). IP-Western analysis was performed using equal amounts of protein as described previously (The et al. 1997). Antibodies to detect ERK (M5670) and p-ERK (M8159) were from Sigma. MEK1 (9122), p-MEK1 (9121), Akt1 (4054), and p-Akt1 (9272) antibodies were from Cell Signaling.
Acknowledgments
We thank Drs. Holger Apitz, Douglas Armstrong, Charles Dearolf, Jay Hirsh, David Hughes, Rob Jackson, Kim Kaiser, Günther Korge, Samuel Kunes, Sally Leevers, Fumio Matsuzaki, Christen Mirth, Michael O’Connor, Eric Rulifson, Susan St. Pierre, Paul Taghert, Stefan Thor, and Scott Waddell for mutants and transgenic lines. We are grateful to Alfred Wittinghofer for advice on GRD mutants, to Doug Rennie for embryo injections, and to Jeffrey Settleman for helpful comments. This work was supported by a Public Health Service grant to I.K.H., by grants from the U.S. Army Medical Research and Materiel Command to A.B., and by Young Investigator Awards by the Children’s Tumor Foundation to S.B., A.V.T., and J.A.W.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1466806
References
- Ahmadian, M.R., Kiel, C., Stege, P., Scheffzek, K. Structural fingerprints of the Ras-GTPase activating proteins neurofibromin and p120GAP. J. Mol. Biol. 2003;329:699–710. doi: 10.1016/s0022-2836(03)00514-x. [DOI] [PubMed] [Google Scholar]
- Almeida, M.S., Bray, S.J. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 2005;122:1282–1293. doi: 10.1016/j.mod.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Aravind, L., Neuwald, A.F., Ponting, C.P. Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr. Biol. 1999;9:R195–R197. doi: 10.1016/s0960-9822(99)80127-4. [DOI] [PubMed] [Google Scholar]
- Beadle, G., Tatum, E., Clancy, C. Food level in relation to rate of development and eye pigmentation in. Drosophila melanogaster. Biol. Bull. 1938;75:447–462. [Google Scholar]
- Bohni, R., Riesgo-Escovar, J., Oldham, S., Brogiolo, W., Stocker, H., Andruss, B.F., Beckingham, K., Hafen, E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- Boyanapalli, M., Lahoud, O.B., Messiaen, L., Kim, B., de Anrle Sylor, M.S., Duckett, S.J., Somara, S., Mikol, D.D. Neurofibromin binds to caveolin-1 and regulates ras, FAK, and Akt. Biochem. Biophys. Res. Commun. 2006;340:1200–1208. doi: 10.1016/j.bbrc.2005.12.129. [DOI] [PubMed] [Google Scholar]
- Brand, A.H., Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brill, S., Li, S., Lyman, C.W., Church, D.M., Wasmuth, J.J., Weissbach, L., Bernards, A., Snijders, A.J. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell. Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, P.E., Walkiewicz, M., Stern, M. Ras activity in the Drosophila prothoracic gland regulates body size and developmental rate via ecdysone release. Curr. Biol. 2005;15:1785–1795. doi: 10.1016/j.cub.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Chen, C., Jack, J., Garofalo, R.S. The Drosophila insulin receptor is required for normal growth. Endocrinology. 1996;137:846–856. doi: 10.1210/endo.137.3.8603594. [DOI] [PubMed] [Google Scholar]
- Cichowski, K., Jacks, T. NF1 tumor suppressor gene function: Narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- Colombani, J., Bianchini, L., Layalle, S., Pondeville, E., Dauphin-Villemant, C., Antoniewski, C., Carre, C., Noselli, S., Leopold, P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- Costa, R.M., Federov, N.B., Kogan, J.H., Murphy, G.G., Stern, J., Ohno, M., Kucherlapati, R., Jacks, T., Silva, A.J. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- D’Angelo, I., Welti, S., Bonneau, F., Scheffzek, K. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep. 2006;7:174–179. doi: 10.1038/sj.embor.7400602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, B., Dugan, L.L., Gutmann, D.H. The neurofibromatosis 1 gene product neurofibromin regulates pituitary adenylate cyclase-activating polypeptide-mediated signaling in astrocytes. J. Neurosci. 2003;23:8949–8954. doi: 10.1523/JNEUROSCI.23-26-08949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, B., Yi, Y., Chen, D.Y., Weber, J.D., Gutmann, D.H. Proteomic analysis reveals hyperactivation of the mammalian target of rapamycin pathway in neurofibromatosis 1-associated human and mouse brain tumors. Cancer Res. 2005;65:2755–2760. doi: 10.1158/0008-5472.CAN-04-4058. [DOI] [PubMed] [Google Scholar]
- Davidowitz, G., D’Amico, L.J., Nijhout, H.F. Critical weight in the development of insect body size. Evol. Dev. 2003;5:188–197. doi: 10.1046/j.1525-142x.2003.03026.x. [DOI] [PubMed] [Google Scholar]
- Fahsold, R., Hoffmeyer, S., Mischung, C., Gille, C., Ehlers, C., Kucukceylan, N., Abdel-Nour, M., Gewies, A., Peters, H., Kaufmann, D., et al. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am. J. Hum. Genet. 2000;66:790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, P., Eicher, E.N., Leevers, S.J., Hafen, E., Hughes, D.A. Control of growth and differentiation by Drosophila RasGAP, a homolog of p120 Ras-GTPase-activating protein. Mol. Cell. Biol. 1999;19:1928–1937. doi: 10.1128/mcb.19.3.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul, U., Mardon, G., Rubin, G.M. A putative Ras GTPase activating protein acts as a negative regulator of signaling by the Sevenless receptor tyrosine kinase. Cell. 1992;68:1007–1019. doi: 10.1016/0092-8674(92)90073-l. [DOI] [PubMed] [Google Scholar]
- Guo, H.F., The, I., Hannan, F., Bernards, A., Zhong, Y. Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science. 1997;276:795–798. doi: 10.1126/science.276.5313.795. [DOI] [PubMed] [Google Scholar]
- Guo, H.F., Tong, J., Hannan, F., Luo, L., Zhong, Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature. 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- Gutmann, D.H., Boguski, M., Marchuk, D., Wigler, M., Collins, F.S., Ballester, R. Analysis of the neurofibromatosis type 1 (NF1) GAP-related domain by site-directed mutagenesis. Oncogene. 1993;8:761–769. [PubMed] [Google Scholar]
- Hannan, F., Ho, I., Tong, J.J., Zhu, Y., Nurnberg, P., Zhong, Y. Effect of neurofibromatosis type I mutations on a novel pathway for adenylyl cyclase activation requiring neurofibromin and Ras. Hum. Mol. Genet. 2006;15:1087–1098. doi: 10.1093/hmg/ddl023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, S.D., Solomon, N., Rubin, G.M. A genetic analysis of the 63E–64A genomic region of Drosophila melanogaster: Identification of mutations in a replication factor C subunit. Genetics. 1995;139:1701–1709. doi: 10.1093/genetics/139.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingtgen, C.M., Roy, S.L., Clapp, D.W. Stimulus-evoked release of neuropeptides is enhanced in sensory neurons from mice with a heterozygous mutation of the Nf1 gene. Neuroscience. 2006;137:637–645. doi: 10.1016/j.neuroscience.2005.09.030. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Rangwala, F., Fulkerson, P.C., Ling, B., Reed, E., Cox, A.D., Kamholz, J., Ratner, N. Role of TC21/R-Ras2 in enhanced migration of neurofibromin-deficient Schwann cells. Oncogene. 2004;23:368–378. doi: 10.1038/sj.onc.1207075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson, S.M., Hughes, R.A.C., editors. The Neurofibromatoses: A pathogenetic and clinical overview. Chapman & Hall Medical; London: 1994. [Google Scholar]
- Ikeya, T., Galic, M., Belawat, P., Nairz, K., Hafen, E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Johannessen, C.M., Reczek, E.E., James, M.F., Brems, H., Legius, E., Cichowski, K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl. Acad. Sci. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, A., Ahmadian, M.R., Schuelke, M., Scheffzek, K., Hoffmeyer, S., Gewies, A., Schmitz, F., Kaufmann, D., Peters, H., Wittinghofer, A., et al. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum. Mol. Genet. 1998;7:1261–1268. doi: 10.1093/hmg/7.8.1261. [DOI] [PubMed] [Google Scholar]
- Kluwe, L., Friedrich, R.E., Peiper, M., Friedman, J., Mautner, V.F. Constitutional NF1 mutations in neurofibromatosis 1 patients with malignant peripheral nerve sheath tumors. Hum. Mutat. 2003;22:420. doi: 10.1002/humu.9193. [DOI] [PubMed] [Google Scholar]
- Leevers, S.J., Weinkove, D., MacDougall, L.K., Hafen, E., Waterfield, M.D. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 1996;15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- Li, H., Chaney, S., Roberts, I.J., Forte, M., Hirsh, J. Ectopic G-protein expression in dopamine and serotonin neurons blocks cocaine sensitization in Drosophila melanogaster. Curr. Biol. 2000;10:211–214. doi: 10.1016/s0960-9822(00)00340-7. [DOI] [PubMed] [Google Scholar]
- Lundell, M.J., Chu-LaGraff, Q., Doe, C.Q., Hirsh, J. The engrailed and huckebein genes are essential for development of serotonin neurons in the Drosophila CNS. Mol. Cell. Neurosci. 1996;7:46–61. doi: 10.1006/mcne.1996.0004. [DOI] [PubMed] [Google Scholar]
- Mattocks, C., Baralle, D., Tarpey, P., Ffrench-Constant, C., Bobrow, M., Whittaker, J. Automated comparative sequence analysis identifies mutations in 89% of NF1 patients and confirms a mutation cluster in exons 11–17 distinct from the GAP related domain. J. Med. Genet. 2004;41:e48. doi: 10.1136/jmg.2003.011890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth, C., Truman, J.W., Riddiford, L.M. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr. Biol. 2005;15:1796–1807. doi: 10.1016/j.cub.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Neufeld, T.P., de la Cruz, A.F., Johnston, L.A., Edgar, B.A. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Ohba, Y., Mochizuki, N., Yamashita, S., Chan, A.M., Schrader, J.W., Hattori, S., Nagashima, K., Matsuda, M. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J. Biol. Chem. 2000;275:20020–20026. doi: 10.1074/jbc.M000981200. [DOI] [PubMed] [Google Scholar]
- Poullet, P., Lin, B., Esson, K., Tamanoi, F. Functional significance of lysine 1423 of neurofibromin and characterization of a second site suppressor which rescues mutations at this residue and suppresses RAS2Val-19-activated phenotypes. Mol. Cell. Biol. 1994;14:815–821. doi: 10.1128/mcb.14.1.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey, I., Taylor-Harris, P., van Erp, H., Hall, A. R-ras interacts with rasGAP, neurofibromin and c-raf but does not regulate cell growth or differentiation. Oncogene. 1994;9:685–692. [PubMed] [Google Scholar]
- Rorth, P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson, E.J., Kim, S.K., Nusse, R. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Salvaterra, P.M., Kitamoto, T. Drosophila cholinergic neurons and processes visualized with Gal4/UAS-GFP. Brain Res. Gene Expr. Patterns. 2001;1:73–82. doi: 10.1016/s1567-133x(01)00011-4. [DOI] [PubMed] [Google Scholar]
- Salzberg, A., Cohen, N., Halachmi, N., Kimchie, Z., Lev, Z. The Drosophila Ras2 and Rop gene pair: A dual homology with a yeast Ras-like gene and a suppressor of its loss-of-function phenotype. Development. 1993;117:1309–1319. doi: 10.1242/dev.117.4.1309. [DOI] [PubMed] [Google Scholar]
- Scheffzek, K., Ahmadian, M.R., Kabsch, W., Wiesmuller, L., Lautwein, A., Schmitz, F., Wittinghofer, A. The Ras–RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Sermon, B.A., Lowe, P.N., Strom, M., Eccleston, J.F. The importance of two conserved arginine residues for catalysis by the ras GTPase-activating protein, neurofibromin. J. Biol. Chem. 1998;273:9480–9485. doi: 10.1074/jbc.273.16.9480. [DOI] [PubMed] [Google Scholar]
- Shingleton, A.W., Das, J., Vinicius, L., Stern, D.L. The temporal requirements for insulin signaling during development in Drosophila. PLoS Biol. 2005;3:e289. doi: 10.1371/journal.pbio.0030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund, T., Korge, G. Innervation of the ring gland of Drosophila melanogaster. J. Comp. Neurol. 2001;431:481–491. doi: 10.1002/1096-9861(20010319)431:4<481::aid-cne1084>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Skoulakis, E.M., Kalderon, D., Davis, R.L. Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron. 1993;11:197–208. doi: 10.1016/0896-6273(93)90178-t. [DOI] [PubMed] [Google Scholar]
- Taghert, P.H., Hewes, R.S., Park, J.H., O’Brien, M.A., Han, M., Peck, M.E. Multiple amidated neuropeptides are required for normal circadian locomotor rhythms in Drosophila. J. Neurosci. 2001;21:6673–6686. doi: 10.1523/JNEUROSCI.21-17-06673.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar, M., Kopelman, A., Epstein, D., Tu, M.P., Yin, C.M., Garofalo, R.S. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. 2001;292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- The, I., Hannigan, G.E., Cowley, G.S., Reginald, S., Zhong, Y., Gusella, J.F., Hariharan, I.K., Bernards, A. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science. 1997;276:791–794. doi: 10.1126/science.276.5313.791. [DOI] [PubMed] [Google Scholar]
- Tong, J., Hannan, F., Zhu, Y., Bernards, A., Zhong, Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat. Neurosci. 2002;5:95–96. doi: 10.1038/nn792. [DOI] [PubMed] [Google Scholar]
- Vandenbroucke, I., Vandesompele, J., De Paepe, A., Messiaen, L. Quantification of NF1 transcripts reveals novel highly expressed splice variants. FEBS Lett. 2002;522:71–76. doi: 10.1016/s0014-5793(02)02887-9. [DOI] [PubMed] [Google Scholar]
- Waddell, S., Armstrong, J.D., Kitamoto, T., Kaiser, K., Quinn, W.G. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Nicol, G.D., Clapp, D.W., Hingtgen, C.M. Sensory neurons from Nf1 haploinsufficient mice exhibit increased excitability. J. Neurophysiol. 2005;94:3670–3676. doi: 10.1152/jn.00489.2005. [DOI] [PubMed] [Google Scholar]
- Weinkove, D., Neufeld, T.P., Twardzik, T., Waterfield, M.D., Leevers, S.J. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr. Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- Williams, J.A., Su, H.S., Bernards, A., Field, J., Sehgal, A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- Wu, R., Legius, E., Robberecht, W., Dumoulin, M., Cassiman, J.J., Fryns, J.P. Neurofibromatosis type I gene mutation in a patient with features of LEOPARD syndrome. Hum. Mutat. 1996;8:51–56. doi: 10.1002/(SICI)1098-1004(1996)8:1<51::AID-HUMU7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Xu, H., Gutmann, D.H. Mutations in the GAP-related domain impair the ability of neurofibromin to associate with microtubules. Brain Res. 1997;759:149–152. doi: 10.1016/s0006-8993(97)00328-4. [DOI] [PubMed] [Google Scholar]
- Yeh, E., Gustafson, K., Boulianne, G.L. Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila. Proc. Natl. Acad. Sci. 1995;92:7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Romero, M.I., Ghosh, P., Ye, Z., Charnay, P., Rushing, E.J., Marth, J.D., Parada, L.F. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes & Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Ghosh, P., Charnay, P., Burns, D.K., Parada, L.F. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]