Abstract
The histone variant mH2A is believed to be involved in transcriptional repression, but how it exerts its function remains elusive. By using chromatin immunoprecipitation and tandem affinity immunopurification of the mH2A1.1 nucleosome complex, we identified numerous genes with promoters containing mH2A1.1 nucleosomes. In particular, the promoters of the inducible Hsp70.1 and Hsp70.2 genes, but not that of the constitutively expressed Hsp70.8, were highly enriched in mH2A1.1. PARP-1 was identified as a part of the mH2A1.1 nucleosome complex and was found to be associated with the Hsp70.1 promoter. A specific interaction between mH2A1.1 and PARP-1 was demonstrated and found to be associated with inactivation of PARP-1 enzymatic activity. Heat shock released both mH2A1.1 and PARP-1 from the Hsp70.1 promoter and activated PARP-1 automodification activity. The data we present point to a novel mechanism for control of Hsp70.1 gene transcription. mH2A1.1 recruits PARP-1 to the promoter, thereby inactivating it. Upon heat shock, the Hsp70.1 promoter-bound PARP-1 is released to activate transcription through ADP-ribosylation of other Hsp70.1 promoter-bound proteins.
Keywords: Histone variant, mH2A, PARP-1, ADP-ribosylation, transcription
The fundamental unit of chromatin, the nucleosome core, is a multisubunit structure consisting of four different histone types (Luger et al. 1997). An important way to control chromatin function is to alter the biochemical make-up of the nucleosome by replacing an individual histone with a histone variant. This enables the structure and function of the nucleosome to be regulated in a very precise way (Malik and Henikoff 2003).
The histone variant macroH2A (mH2A) is an unusual core histone related to canonical H2A. mH2A consists of a histone H2A-like domain fused to a large C-terminal nonhistone region (NHR). The NHR is also termed the macro domain (Pehrson and Fried 1992). Two closely related but distinct mH2A genes, mH2A1 and mH2A2, have been identified (Pehrson and Fuji 1998; Chadwick and Willard 2001; Costanzi and Pehrson 2001). mH2A1 has two splice variants, mH2A1.1 and mH2A1.2 (Pehrson et al. 1997).
A number of studies suggest that mH2A could play an essential role in heterochromatin establishment or maintenance (Costanzi and Pehrson 1998; Rasmussen et al. 2000; Chadwick and Willard 2002; Grigoryev et al. 2004). mH2A has been shown to reside in close proximity to pericentric heterochromatin (Costanzi et al. 2000); it was also found associated with the facultative heterochromatin of the inactive X chromosomes in female mammals (Costanzi and Pehrson 1998). However, mH2A is expressed at similar levels in male and female cells (Rasmussen et al. 1999), suggesting that its function is not restricted to X-chromosome inactivation. In vitro, mH2A was found able to inhibit SWI/SNF nucleosome remodeling and to interfere with initiation of Polymerase II transcription (Angelov et al. 2003; Doyen et al. 2006). These data suggest that mH2A could be involved in transcription repression, but precisely how it could exert this function is still largely unknown.
The macrodomain of mH2A bears strong homology with proteins found in several types of single-stranded RNA (ssRNA) viruses (Pehrson and Fuji 1998). The recent crystallization of AF1521, a protein consisting of a stand-alone macro domain from Archaeoglobus fulgidus, suggests that macro domains, including that found in mH2A, could possess catalytic activity toward ADP-ribosylated substrates (Allen et al. 2003). The macro domain product of the yeast gene YBR022w exhibits ADP-ribose-1″-phosphate (Appr-1″-p)-processing activity (Martzen et al. 1999), indicating that macro domains are phosphoesterases. The presence of macro domains in some poly(ADP-ribose) polymerases (PARP-9 and PARP-14) hints at a functional role in poly-ADP-ribosylation (Allen et al. 2003; Ame et al. 2004).
In this work we show that the promoters of the inducible human HSP-70.1 and HSP-70.2 genes, but not the constitutively expressed HSP-70.8 gene (HSC70), are very highly enriched in mH2A1.1. By using a TAP-ChIP (for tandem-affinity purification and chromatin immunoprecipitation assay) approach we have identified promoters of numerous other genes enriched in macroH2A1.1. We also show that mH2A.1.1 interacts specifically with PARP-1 through its macro domain. We present evidence suggesting that this interaction is crucial for the keeping the HSP-70.1 promoter silenced. Our data point to a novel mechanism for the transcriptional repression of HSP-70.1 through down-regulation of the activity of PARP-1 by mH2A1.1, and they suggest that numerous highly regulated genes could be kept silenced by this novel mechanism.
Results
Identification of Hsp70 as a mH2A1.1 target gene
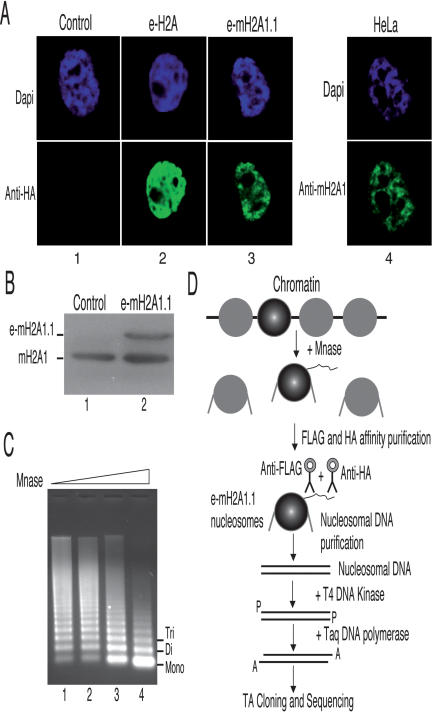
A number of studies point to a general involvement of mH2A in heterochromatin establishment and transcription repression (Costanzi and Pehrson 1998; Angelov et al. 2003; Chadwick and Willard 2004; Grigoryev et al. 2004). The mechanisms by which this repression is achieved are not really known (Angelov et al. 2003; Ladurner 2003; Abbott et al. 2005). We have used a highly specific ChIP assay that is based on the TAP method (Nakatani and Ogryzko 2003) to identify specific DNA sequences associated with mH2A1.1. The approach, termed TAP-ChIP, is schematically described in Figure 1. It uses a HeLa cell line stably expressing mH2A1.1 fused to its N terminus with double HA and Flag peptides (e-mH2A1.1). The immunofluorescence staining of e-mH2A1.1 in these cells reveals that the tagged histones colocalize with chromatin (Fig. 1A), indicating that the presence of the tag epitopes does not interfere with their deposition and association with chromatin. It is important to note that the expression level of the tagged protein did not exceed 30% of the endogenous mH2A1, as evidenced by Western blot using an anti-mH2A1 antibody (Fig. 1B).
Figure 1.
Preparation and purification of e-mH2A1.1 nucleosomes. (A) e-mH2A1.1 associates with chromatin. HeLa cell lines stably expressing either e-H2A (column 2) or e-mH2A1.1 (column 3) were stained with DAPI (top) and anti-HA antibody (bottom). (Column 4) Control HeLa cell line stained with DAPI (top) and anti-macroH2A1 antibody (bottom). (B) Western blot quantification of e-mH2A1.1 expression level in HeLa cells. Total extracts isolated from HeLa cells expressing e-mH2A1.1 or not were resolved by SDS-PAGE, blotted, and revealed with anti-macroH2A1 antibody. (C) Preparation of e-mH2A1.1 nucleosomes. Nuclei from a HeLa cell line stably expressing an epitope-tagged version of mH2A1.1 were digested with increasing amounts of micrococcal nuclease; the DNA from the digested samples was purified and resolved in 1% agarose gel. (D) Schematics of the TAP-ChIP approach used for the purification of e-mH2A1.1 nucleosomes and identification of the DNA sequences associated with e-mH2A1.1.
Nuclei from the tagged cells were digested with controlled amounts of micrococcal nuclease to give predominantly mono- and dinucleosomes (see Fig. 1C, lane 4). Epitope-tagged mH2A1.1 (e-mH2A1.1) nucleosomes were first immunopurified with anti-Flag antibody followed by a second immunopurification with anti-HA antibody. e-mH2A1.1 nucleosomes were proteinase K digested, and the DNA was phenol extracted. The purified DNA was then treated with T4 DNA kinase, and 3′ A-overhangs were added using Taq DNA polymerase (see Fig. 1D). The DNA fragments corresponding to e-mH2A1.1 nucleosomes were cloned into the pcDNA3.1-Topo vector using TA cloning technology. The transformed clones were checked for inserts, sequenced using vector-specific primers, and then a Blast search in the genome database was carried out. This approach has allowed the identification of numerous genes associated with mH2A1.1 (Supplementary Table 1). One of the genomic sequences identified corresponded to the Hsp70.1 gene. Hsp70.1 is inducible in response to heat shock and chemical stress. We further focused on this gene.
mH2A1.1 specifically targets the Hsp70.1 promoter
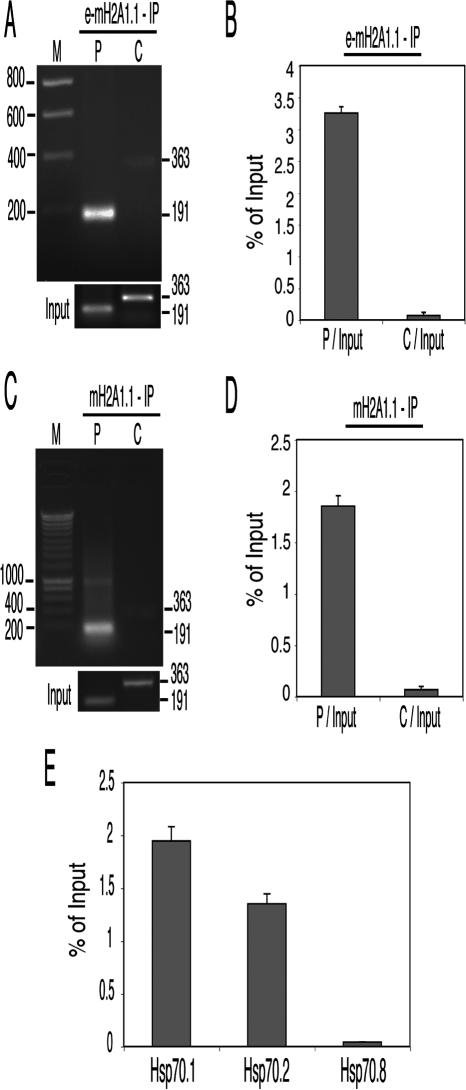
To confirm the association of mH2A1.1 with the Hsp70.1 gene, we have complemented the TAP-ChIP approach with a combination of standard ChIP and either semiquantitative or real-time PCR. The sheared, formaldehyde-treated chromatin from HeLa cells expressing e-mH2A1.1 histone was immunoprecipitated with anti-Flag and anti-HA epitope antibodies. The precipitated chromatin was analyzed using PCR primer pairs that cover either the promoter or the coding region of the Hsp70.1 gene. The PCR products were then run on an agarose gel for analysis. The results show that the DNA fragment spanning the Hsp70.1 promoter (P, 191 base pairs [bp]) was enriched, whereas the fragment located in the coding region (C, 363 bp) was not (see Fig. 2A). We then used real-time PCR to quantify the relative enrichment of e-mH2A1.1 in the promoter region. Two different dilutions of each sample were analyzed independently by Q-PCR with the aforementioned primers. PCR values were normalized against values obtained with chromatin from nontagged cells. The results, presented in Figure 2B, show that e-mH2A1.1 is enriched 45 times on the promoter region of the Hsp70.1 gene compared with the coding region.
Figure 2.
mH2A1.1 is associated with the Hsp70.1 and Hsp70.2, but not with the Hsp70.8 promoter. (A) Analysis of e-mH2A1.1 distribution on the Hsp70.1 locus using semiquantitative PCR. Mono- and dinucleosomes were purified from HeLa cells stably expressing tagged macro-H2A1.1 histone by TAP as described in Figure 1D. Input DNA and DNA isolated from the immunoprecipitated samples were amplified by PCR with Hsp70.1 promoter (P) and coding (C) region primers, and the amplified products were separated on a 2% agarose gel. Molecular masses of the amplification products are indicated on the right side of the figure. Lane M corresponds to a molecular ladder. The panel “Input” (lower part of the figure) shows the fragments amplified from the input fraction (the starting material before immunoprecipitation) with primers specific for the promoter (P) and the coding (C) sequence. (B) e-mH2A1.1 is associated exclusively with the promoter region of the Hsp70.1 gene. Nucleosomes were isolated from a HeLa cell line stably expressing e-mH2A1.1. Input DNA and the DNA isolated from tandem-immunoaffinity-purified e-mH2A1.1 nucleosomes were amplified by real-time PCR. The histograms show the relative enrichment of e-mH2A1.1 on the promoter (P/Input) and the coding region (C/Input) versus the input DNA. (C) Distribution of endogenous mH2A1 on the Hsp70.1 locus using semiquantitative PCR. Nucleosomes were isolated from a control HeLa cell line (not expressing e-mH2A1.1), and the particles associated with the endogenous mH2A1 were immunoprecipitated with an anti-mH2A1 antibody. The input and the DNA isolated from the immunoprecipitated samples were amplified by semiquantitative PCR using the same primers as in A. The amplified products were separated on a 2% agarose gel. Molecular masses of the PCR-amplified fragments are indicated on the right side of the figure. Lane M corresponds to a molecular ladder. At the lower part of the figure (panel “Input”) are shown the fragments amplified from the input material by using primers specific for the promoter (P) and coding (C) sequences. (D) Quantitative real-time PCR of the DNA isolated from the immunoprecipitated nucleosomes with anti-mH2A1 antibody. Nucleosomes were isolated from control HeLa cells and immunoprecipitated with anti-mH2A1 antibody. The DNA isolated from the nucleosomes was amplified by using real-time PCR as described in B. Histograms show the amount of immunoprecipitated DNA as a function of percent input DNA. (E) The promoter regions of the inducible Hsp70.1 and Hsp70.2, but not that of the constitutively expressed Hsp70.8 gene, are highly enriched in mH2A.1. DNA isolated from the nucleosomes immunoprecipitated with the anti-mH2A1 antibody was quantitatively amplified by real-time PCR using primers specific for the promoters of Hsp70.1, Hsp70.2, and Hsp70.8 genes, respectively. Histograms show the amount of DNA immunoprecipitated as a function of percent input DNA.
We next asked if the ectopically expressed mH2A1.1 (e-mH2A1.1) accurately reflects the distribution of native protein throughout the genome and hence if the target genes identified by our TAP-ChIP approach are indeed the real in vivo targets of mH2A1.1. To study this, we examined the in vivo distribution of the endogenous mH2A1 on the Hsp70.1 promoter using a nontagged HeLa cell line. The ChIP assays were performed with a highly specific polyclonal antibody directed against the NHR of mH2A1, and the distribution of mH2A1 on Hsp70.1 was determined by semiquantitative (Fig. 2C) or real-time PCR (Fig. 2D). The results show that the promoter region of Hsp70.1 is enriched 27-fold with endogenous mH2A1 (Fig. 2D).
Note that the antibody used specifically recognizes mH2A1 (it does not react with mH2A2) (data not shown), but it does not differentiate between mH2A1.1 and mH2A1.2, and consequently this ChIP assay cannot really differentiate which of the two splice variants specifically associates with the Hsp70.1 promoter. Nevertheless, bearing in mind that the TAP-ChIP assay identified only mH2A1.1 interacting with the Hsp70.1 promoter, these data could be viewed as confirming the specific association of mH2A1.1 with the Hsp70.1 promoter.
The Hsp70 family encompasses at least 11 different genes. Some of these genes, such as the Hsp70.1 gene, are heat-shock inducible, whereas others are constitutively expressed (Tavaria et al. 1996). We next asked if mH2A1.1 was associated with the promoters of other inducible genes of the Hsp70 family as well as with constitutively expressed Hsp70 genes. We used a candidate approach, focusing on the heat-shock-inducible Hsp70.2 and the constitutively expressed Hsp70.8 (Hsc70) genes (Dworniczak and Mirault 1987). A ChIP assay using the anti-Flag and anti-HA antibodies was carried out with formaldehyde cross-linked chromatin isolated from HeLa cell lines stably expressing e-mH2A1.1. The real-time PCR quantification (using primers specific for the respective promoters) clearly showed that the promoter region of the Hsp70.2 gene was enriched in e-mH2A1.1 to an extent similar to that of the Hsp70.1 gene, whereas the presence of e-mH2A1.1 was barely detectable at the promoter of the Hsp70.8 gene (Fig. 2E). These data suggest that mH2A1.1 may in general be preferentially associated with inducible heat-shock genes.
Characterization of mH2A1.1-associated nucleosomes
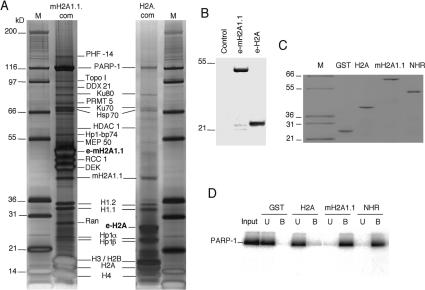
The specific localization of mH2A1.1 on the Hsp70.1 promoter suggests that this histone variant may be associated with the silencing of the promoter of the Hsp70.1 gene. To clarify this question we have identified the proteins specifically associated with mH2A1.1 chromatin. e-mH2A1.1 and e-H2A nucleosomes were purified from the corresponding stably established HeLa cell lines by double immunoaffinity purification using anti-Flag and anti-HA antibodies as described above. The purified complexes were run on 4%–12% gradient polyacrylamide gels containing SDS that were then stained with silver (Fig. 3A). As expected, immunoblotting with anti-Flag antibody revealed one specific band at the expected size for e-mH2A1.1 or e-H2A within the respective complexes (Fig. 3B). Numerous polypeptides were found associated with e-mH2A1.1 and e-H2A nucleosomes (Fig. 3A). Mass spectrometric analyses identified histones H3, H4, H2A, H2B, mH2A1.1, Ku80, Ku70, Hsp70, HDAC1, the two subtypes of H1 (H1.1 and H1.2), HP1α, HP1β, and poly(ADP-ribose) polymerase I (PARP-1) as components common to the e-mH2A1.1 and e-H2A complexes (Fig. 3A). Some proteins, including RCC1, DEK, MEP50, etc, were identified mainly in the mH2A1.1 nucleosome complex. A comprehensive list of the identified proteins, with their accession numbers, is presented in Supplementary Table 2. Although both complexes were found to contain PARP-1, only the e-mH2A1.1 complex had a significant amount of PARP-1. Importantly, the amount of PARP-1 present in the complex was found to be proportional to the amount of e-mH2A1.1 and histone H4 (Fig. 3A; Supplementary Fig. 1), suggesting a direct interaction between mH2A1.1 and PARP-1. In contrast, the H2A complex contained ∼10 times less PARP-1 (the ratio between PARP-1 and histone H4 was found to be close to 0.1) (Fig. 3A, right panel).
Figure 3.
Characterization of H2A and mH2A1.1 complexes. (A) Silver staining of the e-H2A (H2A.com) and e-mH2A1.1 (mH2A1.1.com) nucleosome complexes isolated from HeLa cell lines stably expressing either e-H2A or e-mH2A1.1 proteins. The polypeptides identified by mass spectrometric analyses are indicated. Lane M corresponds to a protein molecular weight marker. The molecular masses of the markers are indicated on the left. (B) Western blot analysis of HeLa cells expressing e-mH2A1.1 and e-H2A. Total extracts isolated from control HeLa cells (control), stable HeLa cells expressing e-mH2A1.1 (e-mH2A1.1), and stable HeLa cells expressing e-H2A (e-H2A) were resolved by SDS-PAGE, blotted, and revealed with anti-Flag antibody. The molecular masses of the protein mass markers are indicated on the left. (C) Purification of recombinant GST-histones. Recombinant GST (GST), GST-H2A (H2A), GST-mH2A1.1 (mH2A1.1), or GST-NHR (NHR) were produced in bacteria and purified on glutathione Sepharose 4B (Amersham) according to the manufacturer’s instructions. The samples were quantified, run on a 12% SDS-PAGE gel, and stained with Coomassie blue. (Lane M) Protein molecular mass marker. (D) GST pull-down experiment. Human recombinant PARP-1 was incubated with either recombinant GST (GST), GST-H2A (H2A), GST-mH2A1.1 (mH2A1.1), or GST-NHR (NHR). Beads were washed, and unbound (U) and bead-bound proteins (B) were run on a 12% SDS-PAGE gel, blotted, and revealed with anti-PARP-1 antibody.
To test if mH2A1.1 interacts specifically with PARP-1, we have expressed and purified fusions of GST with conventional H2A, full-length mH2A1.1, and with its NHR only (Fig. 3C) and have performed a series of GST pull-down experiments (Fig. 3D). A GST-H2A fusion and GST alone were used as controls. Identical amounts of the GST fusions (Fig. 3C), immobilized on glutathione Sepharose beads, were incubated with PARP-1, the beads were washed, and the proteins were eluted from the beads and subjected to SDS-PAGE. After transfer, the blot was revealed with an anti-PARP-1 antibody. Both mH2A1.1 and the NHR were found to interact with PARP-1 (Fig. 3D). This reaction was specific, since only a background signal was observed for GST-H2A and GST (Fig. 3D). These data demonstrate that mH21.1 interacts with PARP-1 through its NHR. Since the Hsp70.1 promoter was highly enriched with mH2A1.1, the above data suggest that the Hsp70.1 should be also enriched with PARP-1 and that the Hsp70.1-associated mH2A1.1 could be involved in the sequestration of PARP-1 on the promoter.
Heat shock induces mH2A1.1 and PARP-1 displacement and massive ADP-ribosylation of the Hsp70.1 promoter-associated proteins
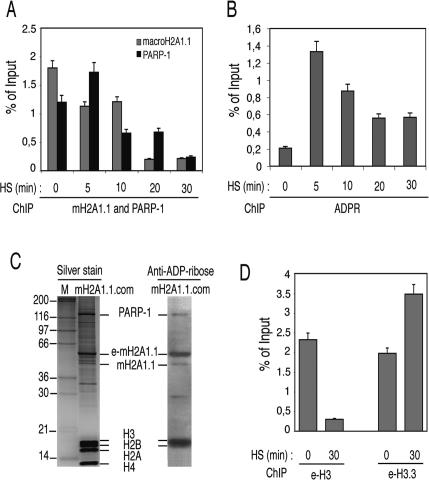
Seeking to comprehensively describe the function of mH2A1.1 in the regulation of Hsp70.1 and its relationship with PARP-1, we have examined the events occurring at the Hsp70.1 promoter during heat-shock-dependent transcriptional activation using a ChIP assay. In addition to the e-mH2A1.1 stable cell line, two other stable HeLa cell lines expressing Flag-HA-tagged versions of either H3 (e-H3) or H3.3 (e-H3.3) histones were established. Initially we focused on the fate of the Hsp70.1 promoter-associated PARP-1 and e-mH2A1.1 upon heat-shock treatment. The cells were heat-shocked for 5, 10, 20, or 30 min at 42°C and, immediately after the heat shock, treated with formaldehyde. Then ChIP was carried out using antibodies against e-mH2A1.1 (anti-Flag or anti-HA antibodies) and against PARP-1, and the amount of bound promoter in the ChIP samples was quantified by Q-PCR (Fig. 4A). As a control, chromatin was immunoprecipitated in the absence of specific antibodies. At 5 min of heat-shock treatment, an ∼30% increase of the amount of PARP-1-associated Hsp70.1 promoter was observed that then sharply decreased upon longer heat-shock treatment, and at 30 min of treatment it already dropped to ∼10% of its initial level before the heat shock. A similar behavior (except at 5 min of heat-shock treatment) was observed for e-mH2A1.1. Analogous ChIP experiments were also carried out using anti-ADP-ribose antibody (Fig. 4B). A background amount of ADP-ribose polymer was present at the Hsp70.1 promoter before heat shock. Five minutes of heat-shock activation resulted in a drastic increase of the promoter-associated ADP-ribose polymer, which decreased upon longer heat-shock treatment (Fig. 4B). Remarkably, the time course of decrease of the ADP-ribose polymer associated with the Hsp70.1 promoter closely followed that of PARP-1 (Fig. 4, cf. A and B). These results provide evidence that macroH2A1.1 and PARP-1 were present on the Hsp70.1 promoter before activation and that heat shock induced their displacement from the promoter. Since ADP-ribosylation is essential for the release of chromatin-bound proteins (for a recent review, see Kim et al. 2005), we hypothesized that both macroH2A1 and PARP-1 could be ADP-ribosylated. To test this, we isolated e-mH2A1.1 nucleosome complexes and used a highly specific anti-ADP-ribose antibody (Supplementary Fig. 5) to visualize the association of the polymer with the proteins of the e-mH2A1.1 nucleosome complex in the absence of heat shock (Fig. 4C). The Western blot showed that both PARP-1 and e-mH2A1.1, as well as the core histones H3 and H2B, were ADP-ribosylated (Fig. 4C, right panel), a result in agreement with the literature (Abbott et al. 2005). These results suggested that upon heat-shock activation not only mH2A1.1 and PARP-1, but also the core histones, should be heavily ribosylated and consequently released from the Hsp70.1 promoter in an ADP-ribosylation-dependent manner. And this was indeed found to be the case, since upon 30 min of heat-shock treatment the amount of histone e-H3-associated Hsp70.1 promoter dropped to ∼10% of its initial value before the heat shock (Fig. 4D, left panel). In parallel, the amount of the histone variant e-H3.3-interacting Hsp70.1 promoter increases close to twofold (Fig. 4D, right panel), a result in agreement with the reported data for other genes (Chow et al. 2005; Mito et al. 2005; Schwartz and Ahmad 2005).
Figure 4.
ChIP analysis reveals a dynamic protein exchange and a dramatic increase of protein ADP-ribosylation of the Hsp70.1 promoter upon heat-shock activation. Distinct HeLa cell lines stably expressing Flag-HA-tagged versions of either mH2A1.1, or H3 or H3.3 histones were used in the experiments. A nontagged HeLa cell line was used as a negative control. (A) Time course of the heat-shock-induced displacement of mH2A1.1 and PARP-1 from the Hsp70.1 promoter. The cells were heat-shocked for the times indicated and then immediately treated with formaldehyde to cross-link the proteins to DNA. ChIP was carried out by using either anti-Flag or anti-PARP-1 antibodies. The DNA isolated from the immunoprecipitated samples was amplified by real-time PCR. The histograms show the amount of Hsp70.1 immunoprecipitated promoter DNA as a percent of the input DNA. The means and standard deviations of three independent experiments are presented. (B) Time course of the ADP-ribosylation of the proteins associated with the Hsp70.1 promoter upon heat shock. The experiment was carried out as in A, but using anti-ADP-ribose antibody. The amounts of ADP-ribose on the Hsp70.1 promoter at the indicated times after the heat shock are presented as copy numbers of the PCR-amplified products. The means and standard deviations of three independent experiments are presented. (C) mH2A1.1 is ADP-ribosylated. (Left panel) The e-mH2A1.1 complex was isolated from stable HeLa cell lines, separated on a 12% PAGE containing SDS, and silver-stained. The first lane shows the molecular markers with the molecular masses indicated at left. The right panel shows the Western blot of the complex revealed by anti-ADP-ribose antibody. Note that e-mH2A1.1, H3, and H2B are heavily ADP-ribosylated. PARP-1 is also found to be ADP-ribosylated, but to a lesser extent. (D) Removal of histone H3 and replacement with H3.3 at the Hsp70.1 promoter upon heat shock. The ChIP experiments were carried out as described in A by using anti-Flag antibody and cells expressing either epitope-tagged H3 (e-H3) or epitope-tagged H3.3 (e-H3.3). The DNA isolated from the immunoprecipitated samples was amplified by real-time PCR. The histograms show the amount of Hsp70.1 promoter DNA immunoprecipitated before (0 min) and after 30 min of heat shock as a percent of input DNA. The means and standard deviations of three independent experiments are given.
These observations suggest that mH2A1.1 could be associated with the establishment and/or maintenance of the inactive state of the Hsp70.1 promoter and that heat-shock activation induces its displacement from the promoter. In addition, our results indicate that PARP-1 ADP-ribosylation activity is inhibited while PARP-1 is sequestered (possibly by mH2A1.1) on the promoter. PARP-1 becomes active when it is released from the Hsp70.1 promoter and modifies promoter-associated histones, thus leading to an accumulation of ADP-ribose moieties.
mH2A1.1 regulates PARP-1 enzymatic activity
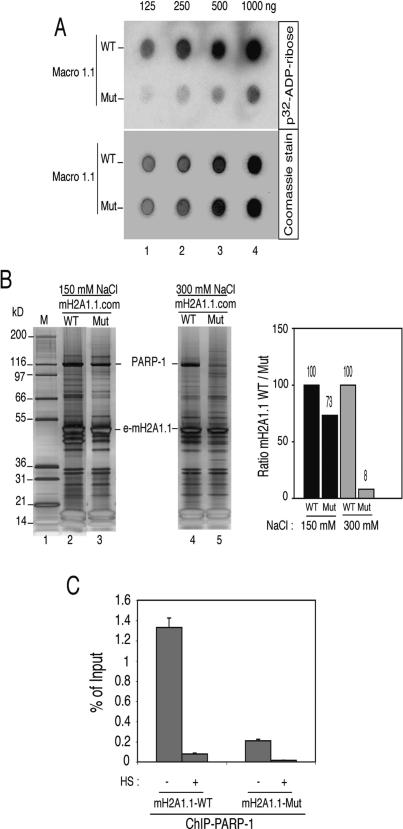
To study the above-suggested interference of mH2A1.1 with the enzymatic activity of PARP-1, we carried out a series of in vitro experiments (Figs. 5 6). The objective was to test if a perturbation in the folding of mH2A1.1 might affect PARP-1 activity and facilitate the release of the otherwise mH2A1.1 nucleosome-associated PARP-1. To this end we generated point mutations in the mH2A1.1 gene that convert a HXTX motif at positions 213–216 in mH2A1.1 to alanines (AAAA) (Supplementary Fig. 4). These mutations were chosen since they could affect a mH2A1.1 putative phosphoesterase activity (Allen et al. 2003). Indeed, this HXTX motif is present in other macro domains that possess catalytic activity toward ADP-ribosylated substrates (Supplementary Fig. 4; Allen et al. 2003). Mutations in this motif abolish the phosphoesterase activity of the macro-domain proteins (Hofmann et al. 2000; Nasr and Filipowicz 2000). This motif is also in close proximity to the mH2A1.1 pocket (Allen et al. 2003; Chakravarthy et al. 2005), which binds ADP ribose (and possibly mono-ADP-ribosylated PARP-1) and its derivative O-acetyl-ADP-ribose (Karras et al. 2005; Kustatscher et al. 2005), so that mutations in the motif may be expected to affect the binding.
Figure 5.
Weaker binding of PARP-1 within the mutated mH2A1.1 nucleosome complex. (A) Altered binding of mono-ADP-ribose to the mutated mH2A1.1. Recombinant wild-type (WT) and mutated (Mut) mH2A1.1 were purified to homogeneity. Increasing amounts of both proteins were loaded (in duplicate) on filters. One filter was then incubated with 32P-ADP-ribose (upper panel), whereas the other one was stained with Coommassie blue as a control for equal loading (lower panel). Note that the binding of the 32P-ADP-ribose to the mutated mH2A1.1 was much weaker compared with that for the wild-type protein. (B) Increasing the ionic strength releases the mutated, but not the wild-type, e-mH2A1.1 protein from the nucleosome complex. The wild-type (WT) and mutated (Mut) mH2A1.1 nucleosome complexes were isolated from cell lines stably expressing the WT or the mutated e-mH2A1.1 histone using either 150 mM NaCl (left panel) or 300 mM NaCl (right panel). The complexes were run on a 4%–12% PAGE gradient containing SDS, then silver-stained. The positions of e-mH2A1.1 and PARP-1 are indicated. (M) Protein molecular mass marker. The right panel presents the quantification of PARP-1 (relative to histone H1) within the WT and mutated e-mH2A1.1 complexes, isolated in 150 mM and 300 mM NaCl, respectively. Note the drastic decrease of the amount of PARP-1 within the mutated mH2A1.1 complex isolated at 300 mM NaCl. (C) The amount of PARP-1 associated in vivo with the Hsp70.1 promoter in the stable cell lines expressing mutated e-mH2A1.1 is much lower compared with that associated with the Hsp70.1 promoter in the stable cell lines expressing WT e-mH2A1.1. Non-heat-shocked (HS; −) and heat-shocked (for 30 min at 42°C) (HS; +) cell lines were treated with formaldehyde to cross-link the proteins to DNA, and ChIP was carried out using anti-PARP-1 antibody. Amounts of the real-time PCR-amplified Hsp70.1 promoter DNA fragments are presented as a percent of input DNA. Note the strong decrease of the amount of PARP-1 associated with the Hsp70.1 promoter of the control cells expressing the mutated e-mH2A1.1, compared with that of the control cells expressing the WT protein.
To test if the above mutations affect the binding of mono-ADP-ribose, increasing amounts of recombinant wild-type (WT) and mutated (Mut) mH2A1.1 macro domains were purified to homogeneity, incubated with 32P-ADP-ribose, and blotted (in duplicate) onto PVDF membranes to detect the labeled proteins (Fig. 5A, upper panel). The duplicate membrane was Coomassie-stained as a control for equal loading (Fig. 5A, lower panel). The obtained results confirmed the weaker binding of mono-ADP-ribose to the mutated e-mH2A1.1 compared with its binding to the WT e-mH2A1.1 (Fig. 5A, upper panel). This provides evidence that the structure of the e-mH2A1.1 pocket that binds ADP-ribose (Karras et al. 2005; Kustatscher et al. 2005) was perturbed in the mutated protein.
If the folding of mH2A1.1 is important for the affinity of PARP-1 binding within the e-mH2A1.1 nucleosome complex, alterations in the folding of e-mH2A1.1 should affect PARP-1 binding. To address this, we have purified WT and Mut e-mH2A1.1 complexes under different conditions of stringency; i.e., at 150 mM (Fig. 5B, left panel) and 300 mM (Fig. 5B, middle panel) NaCl. Remarkably, the amount of PARP-1 associated with the WT and Mut complexes isolated at 150 mM NaCl differed only slightly (see the quantification at Fig. 5B, right panel). However, the picture was completely different for the complexes isolated at 300 mM NaCl (Fig. 5B, middle panel and quantification). In this case, the amount of PARP-1 that remained associated with the mutated e-mH2A1.1 nucleosome complex did not exceed 8%–10% of that associated with the WT e-mH2A1.1 nucleosome complex (Fig. 5B, quantification). Therefore, 300 mM NaCl was able to strongly perturb the binding of PARP-1 to the mutated e-mH2A1.1 within the nucleosome complex, arguing for weaker interaction between PARP-1 and the mutated e-mH2A1.1. This suggests that, in vivo at the Hsp70.1 promoter, the amount of PARP-1 associated with mutated e-mH2A1.1 would be smaller compared with that associated with the WT e-mH2A1.1. We found this indeed to be the case (Fig. 5C). Briefly, we performed ChIP experiments with anti-PARP-1 by using chromatin isolated from HeLa cell lines stably expressing either WT or mutated e-mH2A1.1, and Q-PCR to quantify the amount of PARP-1-associated Hsp70.1 promoter. The quantification showed that no more than 15% of Hsp70.1 promoter was associated with PARP-1 in the cells expressing the mutated e-mH2A1.1, as compared with that found in the cells expressing the WT protein. Taken together, all of the above data demonstrate, both in vitro and in vivo, that the folding of mH2A1.1 is crucial for the binding of PARP-1 to chromatin.
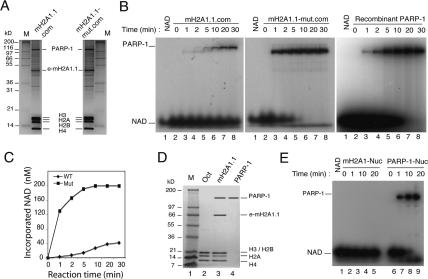
We next measured the auto-ADP-ribosylation activity of the PARP-1 associated with the mutant and wild-type e-mH2A1.1 nucleosome complexes. The two complexes were isolated under the same conditions by double immunoaffinity purification in a buffered solution containing 100 mM NaCl (Fig. 6A). Under these conditions, the relative amount of PARP-1 associated with the mH2A1.1-mut complex was the same as that associated with the wild-type mH2A1.1 complex (Fig. 6A). To measure the auto-ADP-ribosylation activity of PARP-1, wild-type and mutant complexes containing the same amount of associated PARP-1 (100 ng) were incubated in the presence of mononucleosomes and 32P-α-NAD+ (Fig. 6B). A solution containing conventional nucleosomes and recombinant human PARP-1 (100 ng) was used as a positive control (Fig. 6B). The reaction was arrested at the indicated times and the samples loaded on a 12% SDS-PAGE gel. After completion of electrophoresis, the auto-ADP-ribosylated PARP-1 was visualized by autoradiography (Fig. 6B).
Figure 6.
mH2A1.1 down-regulates PARP-1 enzymatic activity. (A) Silver staining of the purified wild-type macroH2A1.1 (e-mH2A1.1) and the mutant (e-mH2A1.1-mut) nucleosomes used for measurement of PARP-1 enzymatic activity. The complexes were isolated using 100 mM NaCl. The bands corresponding to PARP-1, e-mH2A1.1, and the conventional core histones are indicated. (M) Protein molecular mass marker. (B) Kinetic analysis of PARP-1 activity of purified mH2A1.1 complex (left panel), purified mH2A1.1 mutant complex (middle panel), and recombinant PARP-1 (right panel) in the presence of 200 nM 32P-NAD+. To measure the auto-ADP-ribosylation activity of the recombinant PARP-1, an amount of nucleosomes (100 ng) and recombinant PARP-1 equal to that in the mH2A1.1 complexes was used. Note the close auto-ADP-ribosylation kinetics of the recombinant PARP-1 and the PARP-1 in the mH2A1.1-mut.com. (C) Quantification of the data shown in B. Note the dramatic difference in the auto-ADP-ribosylation kinetics of PARP-1 associated with the wild-type and the mutated mH2A1.1 complexes. (D) Twelve percent SDS-PAGE of the purified e-mH2A1.1 octamers and associated PARP-1. e-mH2A1.1 nucleosome complexes were loaded on a hydroxyl apatite column, and after washing with 0.65 M NaCl the remaining proteins were eluted with 2 M buffered solution of NaCl. To purify PARP-1 from the histone octamers, the 2 M NaCl eluate was supplemented with 1 M urea and passed through an agarose-nickel column. (Lanes 1,2) Molecular mass marker and conventional histone octamer as a control. (Lane 3) Tthe protein composition of the 2 M NaCl eluate. (Lane 4) The purified PARP-1. (E) Kinetic analysis of PARP-1 activity associated with in vitro reconstituted nucleosomes containing either conventional core histones (lanes 2–5) or purified e-mH2A1.1 core histones (lanes 6–9). The samples were incubated with 32P-NAD+ and run on 12% PAGE containing SDS. Lane 1 contains 32P-NAD+ only.
Incubation of the wild-type mH2A1.1 complex with mononucleosomes in the presence of 32P-α-NAD+ resulted in a poor labeling of PARP-1 with very slow kinetics (Fig. 6B, left panel). Undeniably, the intensity of the band corresponding to PARP-1 was weak even at 30 min of incubation (the longest time of incubation) (Fig. 6B, left panel). The kinetics and extent of auto-ADP-ribosylation of PARP-1 associated with e-mH2A1.1-mut nucleosomes were, however, completely different (Fig. 6B, middle panel). A strong PARP-1 labeling was already observed at 1 min of incubation; it increased linearly with time and reached completion within 5–10 min (Fig. 6B, middle panel). In addition, the extent of PARP-1 labeling during the first minute was at least 37 times higher than that of the PARP-1 associated with the wild-type e-mH2A1.1 nucleosomes (Fig. 6C, quantification). These results strongly suggest that the interaction of mH2A1.1 with PARP-1 interferes with the PARP-1 auto-ADP-ribosylation activity. Indeed, perturbation of the folding of the mutated mH2A1.1 (Fig. 5A,B) would result in a perturbation of the specific interaction between the mutated mH2A1.1 and PARP-1 (Fig. 5B), which in turn would allow PARP-1 to adopt a conformation close to that of the enzyme free in solution, with higher enzymatic activity. Our data on the auto-ADP-ribosylation of recombinant PARP-1 are in agreement with this, since both the kinetics and the degree of auto-ADP-ribosylation of recombinant PARP-1 were similar to these of the PARP-1 associated with the mutant e-mH2A1.1 (Fig. 6B, cf. middle and right panels). Note that the NAD concentrations used here are close to the physiological concentrations and do not allow the formation of a highly branched PARP-1 (see Supplementary Fig. 2).
Since the e-mH2A1.1 mononucleosome complex contains, in addition to PARP-1, a number of other proteins (Supplementary Table 2), it is difficult to completely exclude the possibility that some of these proteins might, like e-mH2A1.1, interact with PARP-1 and also interfere with its enzymatic activity. To rule this out, we sought to purify the e-mH2A1.1 octamers and the associated PARP-1, to reconstitute the e-mH2A1.1 nucleosome–PARP-1 complex from highly purified components, and to measure the enzymatic activity of the associated e-mH2A1.1 nucleosome PARP-1. To this end, we generated a new HeLa cell line stably expressing a triple-tagged version of mH2A1.1 (Flag-HA-His). This allowed the purification of the e-mH2A1.1 histone octamer and PARP-1 from the e-mH2A1.1 nucleosome complex. Briefly, the Flag-HA purified mononucleosomes were adsorbed on a hydroxyl apatite column and washed with 0.65 M NaCl. Washing the column with 0.65 M NaCl released all of the associated proteins, with the exception of PARP-1, (Fig. 6D, lane 3; data not shown), providing evidence for a very strong binding of PARP-1 to e-mH2A1.1 nucleosomes (note that the e-mH2A1.1 and PARP-1 remaining on the column are in roughly stoichiometric amounts, suggesting that one molecule of nucleosomal e-mH2A1.1 might be complexed with one molecule of PARP-1).
The e-mH2A1.1 octamer and PARP-1 were released from the column with a buffered solution containing 2 M NaCl (Fig. 6D, lane 3) and were used for reconstitution of the e-mH2A1.1–PARP-1 nucleosome complex. The auto-ADP-ribosylation activity of the PARP-1 associated with the reconstituted e-mH2A1.1 nucleosomes was then tested (Fig. 6E). A solution containing conventional nucleosomes and native PARP-1 (purified from the 2 M NaCl eluate of the hydroxyl apatite column-immobilized e mH2A1.1 nucleosome complex) (Fig. 6D, lane 4; see Materials and Methods for details) was used as a positive control (Fig. 6E). The reaction was arrested at the indicated times and the samples were loaded on a 12% SDS-PAGE gel. After completion of electrophoresis, the auto-ADP-ribosylated PARP-1 was visualized by autoradiography (Fig. 6E). Incubation of the in vitro reconstituted e-mH2A1.1 nucleosomes with PARP-1 in the presence of 32P-α-NAD+ resulted in a complete inactivation of its enzymatic activity (Fig. 6E, lanes 2–5), while the same PARP-1, when incubated with conventional mononucleosomes, showed a strong ADP-ribosylation activity, which increased with time and reached completion within 10–20 min (Fig. 6E, lanes 6–9). In addition, incubation of the in vitro reconstituted mH2A1.1 nucleosomes containing PARP-1 in the presence of a 100-fold excess of conventional nucleosomes (Supplementary Fig. 6) failed to reactivate PARP-1, further confirming that the specific interaction between PARP-1 and mH2A1.1 determines the “inactivation” of the enzyme.
Suppression of mH2A1 expression interferes with the heat-shock response
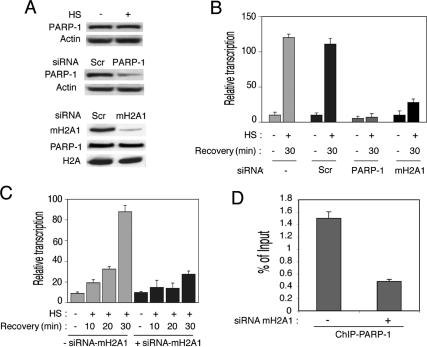
According to reported data, PARP-1 is required for the heat-shock-induced puffing and hsp70 expression in Drosophila larvae (Tulin and Spradling 2003). A whole set of data in this manuscript show that in mammalian cells, PARP-1 is intimately related with mH2A1.1, which in turn suggests that mH2A1.1 should also be involved in the heat-shock response. Bearing this in mind, suppression of the expression of either mH2A1.1 or PARP-1 would be expected to affect the heat-shock response. To test this, we designed two small interfering RNAs (siRNAs) corresponding to the mRNA sequences of either mH2A1 or PARP-1 and used them for transfection in HeLa cells. A scrambled (Scr) sequence was used as a negative control. Forty-eight hours post-transfection, cells were heat-shocked for 30 min at 42°C and left to recover for 30 min at 37°C. Cells were then harvested and assayed both for suppression of the expression of mH2A1.1 and PARP-1 (by Western blotting) and for the transcription of Hsp70.1 (by RT–PCR) (Fig. 7).
Figure 7.
siRNA knockdown of the expression of mH2A1.1 or PARP-1 delays the heat-shock response. (A) The siRNA treatment efficiently suppresses the expression of mH2A1 and PARP-1. HeLa cells were transfected with siRNA specific for either PARP-1 or mH2A.1, or with scrambled siRNA. Forty-eight hours post-transfection the cells were heat-shocked; after a recovery of 30 min, they were collected and lysed. PARP-1 (middle panel) and mH2A.1 (lower panel) were detected by Western blotting using specific antibodies. Immunodetection of actin and H2A was used as control for equal loading. (Lower panel) The level of PARP-1 expression was checked before and after macroH2A1 knock-down. The upper panel shows the amount of PARP-1 before and after heat shock (HS). Note the strong and specific suppression of both PARP-1 and mH2A1 in cells treated with the respective siRNA. (B) Inhibition of mH2A1.1 and PARP-1 expression by siRNA affects the heat-shock response in a similar way. HeLa cells were transfected or not with either macroH2A1-specific siRNA (mH2A1), PARP-1-specific siRNA (PARP-1), or unrelated siRNA (Scr, scrambled). After the heat shock the cells were allowed to recover for 30 min, and RNA was isolated from the different samples. The real-time PCR quantification of the relative amount of Hsp70.1 mRNA versus GAPDH mRNA in HeLa cells before (HS; −) and after (HS; +) a heat shock is shown. Each histogram presents the mean and standard deviation of three independent experiments. (C) Kinetic analysis of Hsp70.1 activation in knocked-down mH2A1.1 HeLa cells. The cells were transfected with mH2A1-specific siRNA (+siRNA-mH2A1) or without any siRNA (−siRNA-mH2A1), and RNA was isolated at 10, 20, or 30 min after the heat shock (HS; +) or in the absence of any heat shock (HS; −). Each histogram is the measured (by real-time PCR) relative amount of Hsp70.1 mRNA versus GAPDH mRNA at the indicated times before (HS; −) or after (HS; +) a heat shock. The means and standard deviations of three independent experiments are presented. (D) Suppression of the expression of mH2A1 with siRNA down-regulates the amount of PARP-1 associated with the Hsp70.1 promoter. Control nontreated (−siRNA-mH2A1) and siRNA-treated (+siRNA-mH2A1) HeLa cells were cross-linked with formaldehyde and used for ChIP with anti-PARP-1 antibody. DNA was isolated from both ChIP samples and submitted to real-time PCR amplification with primers specific for the Hsp70.1 promoter.
Transfection with the specific siRNA strongly inhibited the expression of PARP-1 and mH2A1, whereas transfection with the scrambled control sequence had no effect (Fig. 7A). The RT–PCR revealed that the knockdown of either PARP-1 or mH2A1 dramatically down-regulated transcription of the Hsp70.1 gene, which dropped to background level (the level in the absence of heat shock) in the case of PARP-1 and to about twice the background level when mH2A1 was knocked down (Fig. 7B). In contrast, HeLa cells transfected with a scrambled siRNA (Scr) behaved like the nontransfected control and did not show any repression of Hsp70.1 activation (Fig. 7B).
In order to confirm these results, we also performed a kinetic analysis of Hsp70.1 activation in the presence of macroH2A1 siRNAs (Fig. 7C). HeLa cells transfected or not with macroH2A1 siRNA were heat-shocked for 30 min at 42°C and the expression of Hsp70.1 was monitored by real-time RT–PCR after 10, 20, and 30 min of recovery. The results clearly show that down-regulation of macroH2A1 with a specific siRNA results in a very strong decrease and a delay of the heat-shock response (Fig. 7C). We conclude that the knockdown of mH2A1 has an effect similar to that of PARP-1 knockdown on the heat-shock response. Since mH2A1 appeared to be involved in the association of PARP-1 with the Hsp70.1 promoter, one would expect that the depletion of mH2A1 by siRNA treatment would severely reduce the amount of Hsp70.1 promoter-associated PARP-1, which in turn would interfere with the heat-shock response. The ChIP data (Fig. 7D) demonstrated that the depletion of mH2A1 resulted, as expected, in a strong decrease of the amount of PARP-1 interacting with the Hsp70.1 promoter.
Note that the siRNA used for mH2A1 suppression recognizes both the mRNA of mH2A1.1 and the mRNA of mH2A1.2 (see Materials and Methods for details). Our data thus reflect the effect of the suppression of both mH2A1.1 and mH2A1.2 on the expression of Hsp70.1. However, since only mH2A1.1 was found associated with the Hsp70.1 promoter, we attributed the observed effect only to the ablation of mH2A1.1.
Discussion
We report here a physical interaction between mH2A.1.1 and PARP-1. We present evidence suggesting that this interaction is crucial for keeping the HSP-70.1 promoter silenced through down-regulation of the enzymatic activity of PARP-1 by mH2A1.1. This was confirmed by measuring the PARP-1 auto-ADP-ribosylation activity associated with a nucleosome complex containing a folding mutant of mH2A1.1. Heat-shock activation releases both PARP-1 and mH2A1.1 from the Hsp70.1 promoter and induces a massive ADP-ribosylation of the promoter-associated proteins.
These observations imply that PARP-1 and mH2A1.1 act in concert to regulate the transcriptional status of the Hsp70.1 promoter in mammalian cells. What is the precise mechanism of this regulation? A number of studies have considered PARP-1 as a regulator of chromatin structure and transcription, but conflicting results have left its role unclear (Kraus and Lis 2003; Pirrotta 2004). Several authors have reported a distinct role of PARP-1 in gene regulation (Oei et al. 1998). A recent study reported that Drosophila PARP-1 is a component of a repressive complex, which is also essential for gene activation (Ju et al. 2004). In another study, Drosophila PARP-1 was found to be broadly distributed in the euchromatic arms of polytene chromosomes (Tulin and Spradling 2003). At most sites, PARP-1 is inactive, but its enzymatic activity is required at sites of active transcription such as ecdysteroid and heat-shock-induced puffs (Tulin and Spradling 2003). Therefore, PARP-1 may have a dual function, being in some cases implicated in gene activation, whereas in others it could be implicated in gene silencing. This dual function of PARP-1 appeared to be important since mammalian cells have evolved and express a specialized histone variant mH2A1.1, which through its NHR can sequester PARP-1 to target DNA sequences. Upon association with the mH2A chromatin, PARP-1 becomes a part of a transcriptional repressive complex. Based on our data, we hypothesized that upon heat shock, the folding of the NHR of mH2A1.1 is perturbed. Such perturbation may be the result of a specific NHR modification (e.g., phosphorylation) associated with the heat shock. This perturbation of the folding of mH2A would affect the mH2A1.1-PARP-1 interaction and allow PARP-1 to “recover” its enzymatic activity and to ADP-ribosylate itself and the neighboring histones. The ADP-ribosylation would assist the removal of the histones from the Hsp70.1 promoter and allow the activation of transcription. A massive protein ribosylation would require a dissociation of PARP-1 from mH2A1.1 chromatin, and this is indeed the case, since our ChIP data show no PARP-1 associated with the Hsp70.1 promoter at 30 min of heat shock (Fig. 4). The auto-ADP-ribosylation was not, however, sufficient to release PARP-1 from mH2A1.1 chromatin (see Supplementary Fig. 3), suggesting that another factor, possibly a chromatin remodeling complex, assists in this process.
The above scenario also implies a crucial dual role of mH2A1, in both silencing and activation of transcription of the Hsp70.1 promoter. The silencing, as mentioned above, is achieved through the presence of mH2A1.1 on the Hsp70.1 promoter and the sequestration and subsequent “inactivation” of PARP-1. The role of mH2A1.1 in Hsp70.1 promoter activation involves keeping PARP-1 at the right place (associated with the Hsp70.1 promoter), which upon heat shock and subsequent change in the conformation of mH2A results in having an active enzyme at the Hsp70.1 locus at the right time. Thus, it is not surprising that the knockdown of mH2A1.1, like the knockdown of PARP-1, caused an important decrease and delay in the transcription of the Hsp70.1 gene in response to heat shock (Fig. 7). This mechanism of transcriptional regulation has the advantage of allowing very quick transcriptional responses to environment stimuli.
Material and methods
Plasmid construction
Full-length human cDNA clones of H2A (IMAGE 757500), mH2A1.1 (IMAGE 6187051), H3 (IMAGE 5181234), and H3.3 (IMAGE 4872658) were purchased from Invitrogen. The complete coding sequence from each clone was PCR-amplified with primers incorporating restriction enzyme recognition sites and subcloned into the XhoI–NotI sites of the pREV-HTF retroviral vector or pGEX-5X.1 vector (Amersham) using standard techniques.
Purification of e-H2A and e-mH2A1.1 nucleosome complexes
Nuclear pellets prepared from HeLa cells expressing the H2A and mH2A1.1 proteins fused with N-terminal double-HA and double-Flag epitope tags (e-H2A/e-mH2A1.1) were digested with micrococcal nuclease to give predominantly mononucleosomes. Mononucleosomes containing e-H2A or e-mH2A1.1 were purified from the resulting material by immunoprecipitation on anti-Flag antibody-conjugated agarose. After elution with the Flag peptide, the bound nucleosomes were further affinity-purified by anti-HA antibody-conjugated agarose and eluted with the HA peptide.
GST pull-down assay
GST fusion proteins were expressed in Escherichia coli strain BL21 (pLysS) grown at 25°C. The soluble proteins were purified on glutathione Sepharose 4B beads (Amersham) by standard methods. Human recombinant PARP-1 (Alexis) was incubated for 1 h with the GST fusion proteins at 30°C while gentle mixing in PBS, 100 mM KCl, and 0.02% NP40. Beads were then extensively washed in the same buffer, and the bound proteins were eluted in SDS electrophoresis loading buffer, fractionated on SDS-PAGE, blotted, and revealed by a human anti-PARP-1 antibody (Alexis).
Immunofluorescence
Immunofluorescence was performed using standard procedures. Rat anti-HA antibody (Roche) was used at 1:300 dilution; the secondary antibody used is a goat anti-rat IgG coupled to Alexa Fluor 488 (Molecular Probes) at 1:400 dilution.
RNA isolation and quantification
Total RNA was extracted from 107 HeLa cells and reverse-transcribed using a first-strand cDNA synthesis kit according to the manufacturer’s instructions (Amersham Biosciences). Real-time quantitative PCR was performed using the FastStart DNA Master SYBR Green I kit and the LightCycler apparatus according to the protocol provided by the manufacturer (Roche Molecular Biochemicals). PCR was carried out with the following oligonucleotide pairs: 5′-CAGGTGATCAACGACGGAGACA-3′ and 5′-GTCGATCGTCAGGATGGACACG-3′ for Hsp-70.1, and 5′-GGACCTGACCTGCCGTCTAGAA-3′ and 5′-GGTGT CGCTGTTGAAGTCAGAG-3′ for GAPDH. The signals were normalized to the GAPDH signal. PCR products were verified on a 1.5% agarose gel.
Formaldehyde cross-linking and ChIP
Chromatin was prepared as described in Ferreira et al. (2001), with a cross-linking time of 10 min at 37°C and sonication to an average length of 400–800 bp. ChIPs were performed using the following antibodies: M2 anti-Flag agarose beads (Sigma), anti-PARP-1, and anti-ADP-ribose (Alexis). Samples were analyzed by Q-PCR on a LightCycler, and copy numbers were calculated as described in Ferreira et al. (2001). Two different dilutions of each sample were analyzed independently, and the results from Q-PCR were expressed as the fraction of the total number of input copies that were detected in each immunoprecipitate. Primers were Hsp70.1 promoter: forward, 5′-GGCGAAACCC CTGGAATATTCCCGA-3′, and reverse, 5′-AGCCTTGGGA CAACGGGAG-3′; Hsp70.1 coding region: forward, 5′-CAGGT GATCAACGACGGAGACA-3′, and reverse, 5′-GTCGATCGT CAGGATGGACACG-3′; GAPDH: forward, 5′-GGACCTGA CCTGCCGTCTAGAA-3′, and reverse, 5′-GGTGTCGCTGTT GAAGTCAGAG-3′; Hsp70.2 promoter: forward, 5′-GGCCGA GAGTCAGGGAGGAACC-3′, and reverse, 5′-ACTCTTCCA GCTCCACCACAG-3′; Hsp70.8 promoter: forward, 5′-TGTGG CTTCCTTCGTTATTGGA-3′, and reverse, 5′-AAATACCGC TGCCATCCCACCG-3′.
SiRNA-mediated silencing
HeLa cells in exponential growth were seeded onto six-well plates and transfected with 1 μg of mH2A1 siRNA (5′AAGC AGGGUGAAGUCAGUAA-3′), PARP-1 siRNA (5′-AAGCCU CCGCUCCUGAACAAU-3′) or scrambled control siRNA (5′-CAUGUCAUGUUCACAUCUCTT-3′) using Lipofectamine (Invitrogen). Forty-eight hours post-transfection, cells were heat-shocked for 30 min at 42°C and left to recover for 10, 20, and 30 min at 37°C. Cells were then harvested and assayed for Hsp70.1 expression by RT–PCR and, for mH2A1 and PARP-1 silencing, by immunoblotting. Synthetic siRNAs were purchased from Genset Proligo.
Antibodies
Antibodies employed were as follows: monoclonal anti-Flag antibody M2 (Sigma), monoclonal anti-HA antibody 9E (Roche), monoclonal anti-PARP-1 (Alexis), monoclonal anti-ADP-ribose (Alexis), polyclonal anti-mH2A1 (Stefan Dimitrov, INSERM, Grenoble, France), and anti-α-tubulin antibody (Sigma).
Purification of e-mH2A1.1 octamers and associated PARP-1
e-mH2A1.1 nucleosomes were purified from a HeLa cell line stably expressing a triple-tagged version of mH2A1.1 (Flag-HA-His). The purified mononucleosomes were adsorbed on a hydroxyapatite column and washed with 0.65 M NaCl, and the remaining proteins were eluted with 2 M NaCl. PARP-1 was then dissociated from e-mH2A1.1 octamers by incubation in 2 M NaCl and 1 M urea, and purified from the remaining histones by removing the histidine-tagged mH2A1.1 on an agarose-nickel column. The purified e-mH2A1.1 octamers containing PARP-1 and the free PARP-1 were quantified and frozen at −80°C until use.
Poly(ADP-ribosyl)ation assay
Poly(ADP-ribosyl)ation was performed in vitro in a 20-μL reaction mixture containing 20 mM Tris-HCl (pH 7.8), 50 mM NaCl, 3 mM MgCl2, 0.5 mM DTT, 200 nM α-[32P]NAD (10 μCi/nmol) (Amersham), 100 ng of mononucleosomes, and 100 ng of recombinant PARP-1 or native mH2A1.1-associated PARP-1. The reaction was incubated for 1–30 min at 37°C, stopped with 1% SDS, and directly loaded on a 12% SDS-PAGE gel.
Acknowledgments
We thank C. Cremisi for her continuous support and L.L. Pritchard for critical reading of the manuscript. This work was supported by the CNRS, the Association pour la Recherche sur le Cancer (ARC), the Ligue Nationale contre le Cancer, and by grants from the Ministère de la Recherche (ACI Jeune Chercheur, ACI Biologie Cellulaire Moléculaire et Structurale, BCM0070 and ACI Interface Physique-Chimie-Biologie: DRAB, 2004, #04 2 136) and ANR Project no. NT05-1_41978.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.396106
References
- Abbott, D.W., Chadwick, B.P., Thambirajah, A.A., Ausio, J. Beyond the Xi: MacroH2A chromatin distribution and post-translational modification in an avian system. J. Biol. Chem. 2005;280:16437–16445. doi: 10.1074/jbc.M500170200. [DOI] [PubMed] [Google Scholar]
- Allen, M.D., Buckle, A.M., Cordell, S.C., Lowe, J., Bycroft, M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003;330:503–511. doi: 10.1016/s0022-2836(03)00473-x. [DOI] [PubMed] [Google Scholar]
- Ame, J.C., Spenlehauer, C., de Murcia, G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Angelov, D., Molla, A., Perche, P.Y., Hans, F., Cote, J., Khochbin, S., Bouvet, P., Dimitrov, S. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell. 2003;11:1033–1041. doi: 10.1016/s1097-2765(03)00100-x. [DOI] [PubMed] [Google Scholar]
- Chadwick, B.P., Willard, H.F. Histone H2A variants and the inactive X chromosome: Identification of a second macroH2A variant. Hum. Mol. Genet. 2001;10:1101–1113. doi: 10.1093/hmg/10.10.1101. [DOI] [PubMed] [Google Scholar]
- Chadwick, B.P., Willard, H.F. Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome. J. Cell Biol. 2002;157:1113–1123. doi: 10.1083/jcb.200112074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick, B.P., Willard, H.F. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl. Acad. Sci. 2004;101:17450–17455. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy, S., Gundimella, S.K., Caron, C., Perche, P.Y., Pehrson, J.R., Khochbin, S., Luger, K. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, C.M., Georgiou, A., Szutorisz, H., Maia e Silva, A., Pombo, A., Barahona, I., Dargelos, E., Canzonetta, C., Dillon, N. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi, C., Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Costanzi, C., Pehrson, J.R. MACROH2A2, a new member of the MARCOH2A core histone family. J. Biol. Chem. 2001;276:21776–21784. doi: 10.1074/jbc.M010919200. [DOI] [PubMed] [Google Scholar]
- Costanzi, C., Stein, P., Worrad, D.M., Schultz, R.M., Pehrson, J.R. Histone macroH2A1 is concentrated in the inactive X chromosome of female preimplantation mouse embryos. Development. 2000;127:2283–2289. doi: 10.1242/dev.127.11.2283. [DOI] [PubMed] [Google Scholar]
- Doyen, C.M., An, W., Angelov, D., Bondarenko, V., Mietton, F., Studitsky, V.M., Hamiche, A., Roeder, R.G., Bouvet, P., Dimitrov, S. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 2006;26:1156–1164. doi: 10.1128/MCB.26.3.1156-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworniczak, B., Mirault, M.E. Structure and expression of a human gene coding for a 71 kd heat shock ‘cognate’ protein. Nucleic Acids Res. 1987;15:5181–5197. doi: 10.1093/nar/15.13.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, R., Naguibneva, I., Mathieu, M., Ait-Si-Ali, S., Robin, P., Pritchard, L.L., Harel-Bellan, A. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2001;2:794–799. doi: 10.1093/embo-reports/kve173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev, S.A., Nikitina, T., Pehrson, J.R., Singh, P.B., Woodcock, C.L. Dynamic relocation of epigenetic chromatin markers reveals an active role of constitutive heterochromatin in the transition from proliferation to quiescence. J. Cell Sci. 2004;117:6153–6162. doi: 10.1242/jcs.01537. [DOI] [PubMed] [Google Scholar]
- Hofmann, A., Zdanov, A., Genschik, P., Ruvinov, S., Filipowicz, W., Wlodawer, A. Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction. EMBO J. 2000;19:6207–6217. doi: 10.1093/emboj/19.22.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, B.G., Solum, D., Song, E.J., Lee, K.J., Rose, D.W., Glass, C.K., Rosenfeld, M.G. Activating the PARP-1 sensor component of the groucho/TLE1 corepressor complex mediates a CaMKinase IIδ-dependent neurogenic gene activation pathway. Cell. 2004;119:815–829. doi: 10.1016/j.cell.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Karras, G.I., Kustatscher, G., Buhecha, H.R., Allen, M.D., Pugieux, C., Sait, F., Bycroft, M., Ladurner, A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.Y., Zhang, T., Kraus, W.L. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes & Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kraus, W.L., Lis, J.T. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Kustatscher, G., Hothorn, M., Pugieux, C., Scheffzek, K., Ladurner, A.G. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Ladurner, A.G. Inactivating chromosomes: A macro domain that minimizes transcription. Mol. Cell. 2003;12:1–3. doi: 10.1016/s1097-2765(03)00284-3. [DOI] [PubMed] [Google Scholar]
- Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F., Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Malik, H.S., Henikoff, S. Phylogenomics of the nucleosome. Nat. Struct. Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- Martzen, M.R., McCraith, S.M., Spinelli, S.L., Torres, F.M., Fields, S., Grayhack, E.J., Phizicky, E.M. A biochemical genomics approach for identifying genes by the activity of their products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- Mito, Y., Henikoff, J.G., Henikoff, S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1024–1025. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Nakatani, Y., Ogryzko, V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Nasr, F., Filipowicz, W. Characterization of the Saccharomyces cerevisiae cyclic nucleotide phosphodiesterase involved in the metabolism of ADP-ribose 1″,2″-cyclic phosphate. Nucleic Acids Res. 2000;28:1676–1683. doi: 10.1093/nar/28.8.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oei, S.L., Griesenbeck, J., Ziegler, M., Schweiger, M. A novel function of poly(ADP-ribosyl)ation: Silencing of RNA polymerase II-dependent transcription. Biochemistry. 1998;37:1465–1469. doi: 10.1021/bi9727390. [DOI] [PubMed] [Google Scholar]
- Pehrson, J.R., Fried, V.A. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Pehrson, J.R., Fuji, R.N. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson, J.R., Costanzi, C., Dharia, C. Developmental and tissue expression patterns of histone macroH2A1 subtypes. J. Cell. Biochem. 1997;65:107–113. doi: 10.1002/(sici)1097-4644(199704)65:1<107::aid-jcb11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Pirrotta, V. The ways of PARP. Cell. 2004;119:735–736. doi: 10.1016/j.cell.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Rasmussen, T.P., Huang, T., Mastrangelo, M.A., Loring, J., Panning, B., Jaenisch, R. Messenger RNAs encoding mouse histone macroH2A1 isoforms are expressed at similar levels in male and female cells and result from alternative splicing. Nucleic Acids Res. 1999;27:3685–3689. doi: 10.1093/nar/27.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, T.P., Mastrangelo, M.A., Eden, A., Pehrson, J.R., Jaenisch, R. Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J. Cell Biol. 2000;150:1189–1198. doi: 10.1083/jcb.150.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, B.E., Ahmad, K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes & Dev. 2005;19:804–814. doi: 10.1101/gad.1259805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaria, M., Gabriele, T., Kola, I., Anderson, R.L. A hitchhiker’s guide to the human Hsp70 family. Cell Stress Chaperones. 1996;1:23–28. doi: 10.1379/1466-1268(1996)001<0023:ahsgtt>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin, A., Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]