Abstract
The emergence of chloroquine-resistant Plasmodium falciparum malaria imperils the lives of millions of people in Africa, Southeast Asia and South America. Chloroquine resistance is associated with mutations in the Plasmodium falciparum chloroquine resistance transporter (PfCRT). We expressed chloroquine sensitive (HB3) and resistant (Dd2) pfcrt alleles in HEK293 human embryonic kidney cells. PfCRT localized to the lysosomal limiting membrane and was not detected in the plasma membrane. We observed significant acidification of lysosomes containing PfCRT HB3 and Dd2, with Dd2 acidifying significantly more than HB3. A mutant HB3 allele expressing the K76T mutation (earlier found to be key for chloroquine resistance) acidified to the same extent as Dd2, whereas the acidification of a Dd2 allele expressing the T76K “back mutation” was significantly less than Dd2. Thus, the amino acid at position 76 is both an important determinant of chloroquine resistance in parasites and of lysosomal acidification following heterologous expression. PfCRT may be capable of modulating the pH of the parasite digestive vacuole, and thus chloroquine availability. Chloroquine accumulation and glycyl-phenylalanine-2-naphthylamide-induced release of lysosomal Ca2+ stores were unaffected by PfCRT expression. Cytoplasmic domain mutations did not alter PfCRT sorting to the lysosomal membrane. This heterologous expression system will be useful to characterize PfCRT protein structure and function, and elucidate its molecular role in chloroquine resistance.
Keywords: malaria, PfCRT, acidification, lysosome, chloroquine resistance
INTRODUCTION
Plasmodium falciparum, a blood-borne protozoan parasite, causes over 550 million cases of malaria per year resulting in over 2 million deaths per year, mostly of children under the age of 5 and pregnant woman [1]. For more than 50 years chloroquine (CQ) was a frontline therapy, but resistant strains have emerged worldwide [2, 3]. The development of affordable alternatives to CQ has been slow [4]. The analysis of a genetic cross facilitated the identification of a molecular determinant of P. falciparum CQ resistance [5]. Mutations in this gene, named the P. falciparum chloroquine resistance transporter (pfcrt), are sufficient to confer CQ resistance [6]. This gene encodes a 424 amino acid integral membrane protein, PfCRT, that is present in the membrane of the parasite's digestive vacuole (DV) and is predicted to have ten transmembrane segments. PfCRT displays weak primary sequence similarity to bacterial drug metabolite transporters [7, 8], and has been implicated in the development of resistance to other antimalarial agents, including halofantrine and amantidine [9]. CQ-resistant (CQR) patient isolates contain a K76T mutation in a background that contains several other mutations [5]. Expression of the “back mutation” of T76K in a CQ-resistant allele increases the CQ susceptibility [10]. The endogenous function of PfCRT and the mechanism by which it confers CQ resistance is a subject of intense investigation [see 11, for review].
CQ is a weak base that presumably enters the DV by a solubility-diffusion mechanism where it becomes protonated and trapped in this low pH compartment. CQ binds to one or more forms of ferriprotoporphyrin IX (FPIX), a toxic heme metabolite formed in the parasite's DV, and prevents its crystallization into insoluble hemozoin [12]. This leads to parasite death by an as yet undefined mechanism. CQ binding to FPIX, however, makes it difficult to determine the concentration of free CQ in the DV of CQ resistant and sensitive parasites, which complicates the analysis of CQ uptake data. CQR parasites are reported to have lower DV CQ concentrations [13], although the DV volume is usually not directly measured complicating the calculation of DV CQ concentration. CQ resistance may be conferred by reducing access of CQ to FPIX [14, 15]. Alternatively, CQ resistance may involve an energy-dependent, efflux process [16, 17]. Expression of CQR pfcrt in Dictyostelium reduces CQ uptake compared to cells expressing CQ-sensitive (CQS) pfcrt isoforms [18]. Furthermore, several studies have provided evidence that DV pH is altered in CQR strains, and this may be related to pfcrt mutations and CQ resistance [see 19 for review, 20].
Elucidating the molecular basis for PfCRT's involvement in CQ resistance will be vital for developing improved cost-effective drugs to combat CQR malaria. For example, while verapamil, a calcium channel antagonist, reverses CQ resistance [21], the mechanism of reversal is uncertain, despite evidence that PfCRT is involved [5, 10, 20]. Physiological and biochemical studies of Plasmodium proteins in situ are time consuming and subject to technical barriers [22]. Heterologous expression offers a more convenient system to study PfCRT and to investigate its interactions with drugs [18, 23-25]. Here, we demonstrate robust expression of full length PfCRT in human embryonic kidney (HEK293) cells. We determined that this protein is sorted to the lysosome, an acidic organelle functionally analogous to the acidic DV in P. falciparum. Lysosomal pH was significantly altered in cells transfected with CQS (HB3) or CQR (Dd2) alleles, as well as alleles mutated at the 76 position (HB3-T76K and Dd2-K76T). Expression of CQS and CQR PfCRT isoforms did not affect CQ uptake into HEK293 cells or alter steady-state Ca2+ concentrations within the lysosomes. PfCRT sorting to the lysosome was not substantially altered by a series of cytoplasmic domain mutants, indicating a non-conventional lysosomal targeting mechanism for PfCRT in HEK293 cells.
MATERIALS AND METHODS
Plasmid DNA constructs
Yeast codon-normalized pfcrt cDNA sequences (HB3PfCRT-bad and Dd2PfCRT-bad), generated in the laboratory of Dr. P. Roepe, have been previously reported [23]. These coding sequences were excised from the pGEMHE vector by EcoRI and HindIII digestion and ligated into the pcDNA3.1(+) multiple cloning site (Invitrogen, CA). The biotin acceptor domain (bad) sequences were excised by simultaneous NotI and HindIII digestion and re-ligated to form the HB3 and Dd2 alleles. Both constructs were sequenced in their entirety to ensure that their inferred amino acid sequences corresponded to the published HB3 and Dd2 sequences [5]. To generate monomeric Red Fluorescent Protein (RFP) fusion constructs with PfCRT, the HB3-RFP and Dd2-RFP fusions, an AgeI site was introduced before the stop codon of HB3 and Dd2 using a Quikchange kit (Stratagene, CA). PCR amplification of mRFP1 cDNA from PRSET/mRFP1 [26] incorporated in-frame AgeI and NotI sites from the primers. Digestion with AgeI and NotI and ligation into the digested AgeI/NotI-mutant HB3 or Dd2 plasmid resulted in constructs coding for the fusion proteins. PfCRT constructs with C-terminal myc epitope tags, HB3-myc and Dd2-myc, were generated by ligating in a synthetic double-stranded oligonucleotide coding for an in-frame myc tag with the 3f AgeI/NotI sites. Constructs that had residues 5 to 55 deleted from the N-terminal domain, HB3-NTD-myc and Dd2-NTD-myc, were created using PCR with a looping primer that removed these residues. PCR products and parent plasmids were digested with EcoRI and BsrGI and re-ligated. To make the C-terminal deletion, HB3-CTD-myc, a looping primer was also used, which removed the C-terminal 20 residues from HB3-myc, leaving the C-terminal myc epitope tag intact. The construct lacking both N- and C-terminal cytoplasmic segments, HB3-NTD-CTD-myc, was created by ligating together NotI/HindIII fragments of the single deletion mutants described above. A construct where all of the putative cytoplasmic lysines were mutated to arginine, HB3-kminus, was made by multiple cycles of oligonucleotide-directed mutagenesis of HB3-NTD-CTD-myc using a modified Quikchange protocol [27]. The mutations introduced were: K53R, K56R, K115R, K116R, K120R, K236R, K237R, K239R, K339R, K401R and K402R. Quikchange mutagenesis was also used to create the mutants HB3-RFP-K76T and Dd2-RFP-T76K. All constructs were sequenced at the Albert Einstein College of Medicine DNA Sequencing Facility to verify the correct mutations and to ensure that there were no secondary mutations. The cDNAs for GFP tagged hSNX1 and hSNX4 were a gift from Dr. Corinne Leprince (Institut Curie, Paris, France). GFP-tagged LC3 cDNA was a gift from Dr. Ana Maria Cuervo (Albert Einstein College of Medicine). GFP-tagged wild-type human VPS4 and the mutant hVPS4(EQ) cDNAs were a gift from Dr. Philip Woodman (University of Manchester, UK). CD63 and CD63-AEVM cDNAs were a gift from Dr. Michael Caplan (Yale University School of Medicine).
Cell culture and transient transfection
HEK293 cells stably expressing SV40 large T antigen (293T, American Type Culture Collection) were maintained at 37 °C, 5% CO2 in DMEM supplemented with 10% fetal calf serum, 50 U/ml penicillin/streptomycin and GlutaMAX™-I (2 mM, Invitrogen). pfcrt constructs were transfected by the calcium phosphate method [28]. Briefly, 5 μg DNA in 250 mM CaCl2 was mixed dropwise with 2x HBS (280 mM NaCl, 50 mM HEPES and 1.5 mM NaH2PO4). The precipitate was added directly to a 100 mm dish of cells at 50-70% confluence. The cells were replated the following day at 10% dilution onto 35 mm glass-bottomed dishes (MatTek, MA) for immunofluorescence experiments, 100 mm dishes for Western blotting, or 50 mm dishes containing poly-lysine-coated glass coverslips for physiological recordings. Experiments were performed 48-72 hrs post-transfection. As time from transfection increased, an increasing percentage of transfected cells rounded up but excluded Trypan Blue. This was not observed in control transfections.
Visualization of PfCRT expression by indirect immunofluorescence
Transfected cells grown on 35 mm dishes were washed once with PBS and fixed for 10 min with 4% paraformaldehyde freshly prepared in PBS. After three brief washes in PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min, washed again, then blocked for 20 min with 3% BSA in PBS. After an additional PBS wash, cells were covered with 1 ml of antibody or streptavidin-Cy3 (Molecular Probes, OR) diluted in PBS containing 3% BSA and 0.2% Triton X-100. Cells were incubated for 1 hr at room temperature (RT) with slow agitation with primary antibodies, and for 30 min each with secondary and tertiary reagents, followed by 4 washes in PBS of 5 min each. Selective plasma membrane permeabilization was achieved using the method of Kyttala and coworkers [29]. After the final wash, PBS was replaced by mounting medium (Sigma, MO) and sealed under a plastic coverslip. Fluorescence was visualized on a TE2000U Nikon inverted epifluorescence microscope using 40x and 100x S Fluor oil-immersion objectives (Nikon, Japan). Samples were illuminated using a DeltaRAM V monochromator (Photon Technology International, NJ) with 4 nm bandpass excitation at the selected wavelengths (Cy3, 550 nm; mRFP1, 555 nm; Alexa Fluor 488, 488 nm). Green and red emitted light was collected via standard dichroic mirrors and filter sets for FITC and tetramethylrhodamine respectively (Chroma Technology, NH), and detected with a Coolsnapfx CCD monochrome digital camera at 1.3 megapixel resolution (Roper Scientific, NJ). Images were acquired and exported using either ImageMaster 2.0 software (PTI, NJ) or Imaging Workbench 5.1 (INDEC Biosystems, CA). ImageJ (v1.31, NIH, MD) was used for further image processing, which consisted of background subtraction, contrast enhancement and intensity normalization. For overlay panels, monochrome images from red and green channels were combined in Paintshop Pro 8 (JASC software, MN) to give false-color overlays.
Antibodies
PfCRT protein was detected using a rabbit polyclonal antiserum [5]. The mouse monoclonal antibody to human LAMP-2 was clone H4B4 (Developmental Studies Hybridoma Bank, Iowa), to GM130 was mAb 35 (BD Biosciences, San Jose, CA), and to human calnexin was AF18 (Novus Biologicals, Littleton, CO). Rabbit polyclonal and mouse monoclonal anti-myc antibodies were from AbCam (Cambridge, UK). Mouse monoclonal antibody to cathepsin D was from Sigma, MO. Monoclonal antibodies were detected using goat anti-mouse secondary antibodies conjugated to Alexa Fluor 488 or Cy3 (Molecular Probes, OR). Rabbit polyclonal antibodies were detected via a biotin-conjugated goat-anti rabbit secondary and Cy3-streptavidin (Sigma, MO). Human tubulin was detected directly via a Cy3-conjugated primary antibody (Sigma, MO).
Subcellular fractionation
One 100 mm plate of transfected cells was washed once with PBS, scraped into 2 ml PBS containing a wide-spectrum protease inhibitor cocktail (Pierce, IL), and frozen at −-80 °C. The cell suspension was thawed and triturated 10 times through a 26 gauge needle. The homogenate was centrifuged at 1,500 rpm in a tabletop microfuge at 4 °C to give the nuclear pellet, P1. The supernatant was re-centrifuged at 14,000 rpm in the same microfuge, resulting in the P2 pellet that contained mostly lysosomes and mitochondria. The supernatant was subjected to ultracentrifugation at 45,000 rpm for 1 hr at 4 °C in a Ti-70 rotor (Beckman, CA). The resulting microsomal pellet, P3, was carefully separated from the supernatant, S1, representing the cytoplasm. Each pellet was resuspended in 100 μl of PBS plus protease inhibitors. Protein concentrations were determined spectrophotometrically using the Bradford reagent (BioRad, CA) with BSA as the standard. SDS-PAGE analysis of the cell fractions was performed using 4-20% acrylamide/Tris-HCl ReadiGels (Biorad, CA). For each lane 15 μg of protein was incubated at RT for 10 min in reducing SDS sample buffer for loading. After electrophoresis, proteins were transferred to pre-wetted PVDF membranes (BioRad, CA) for 120 min at 200 mA. Membranes were blocked with 5% dry-milk powder in PBS for 30 min at RT, followed by a brief PBS wash, then incubated with rabbit polyclonal anti-PfCRT antiserum (1:400) or mouse monoclonal anti-myc antibody (1:1000) in blocking solution for one hr at RT. After 4 washes of 5 min each in PBS, secondary antibody (HRP-labeled goat anti-rabbit or anti-mouse, Pierce, IL) was added to the blot for 30 min at a 1:5000 dilution in blocking solution. The blot was washed again four times and developed using enhanced chemiluminescence substrates (Supersignal West Dura, Pierce). Emitted light was collected for 5 – 45 min using a digital camera, and blot images were processed using Fluorchem software (Alpha Innotech Corp, CA).
Measurement of lysosomal pH
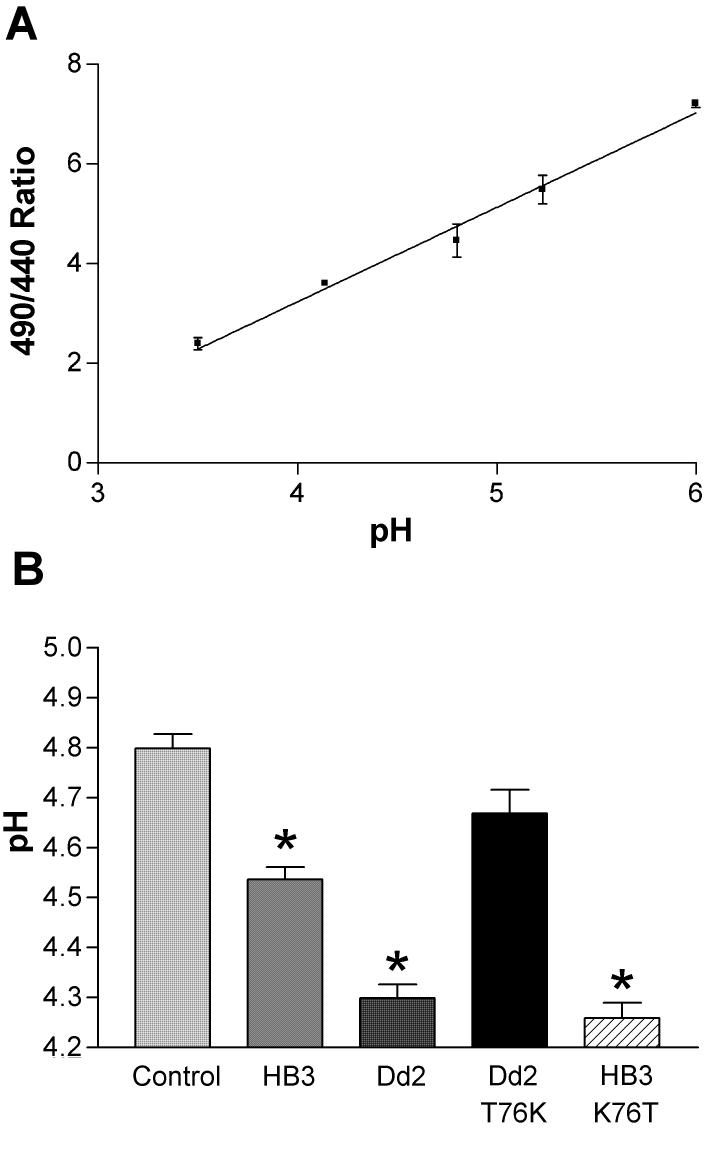
Transfected cells seeded onto 35 mm dishes were incubated in media (as above) for 12 hrs, then incubated in media containing 1 mg/ml BSA and 0.1 mg/ml Oregon Green 488-dextran (Molecular Probes, OR). Loading was continued for 12-18 hrs, before washing once with PBS and chasing with dextran-free media for 6-12 hrs. Oregon Green 488-dextran was visualized under both live imaging and with fixed and permeabilized cells as above, and could be overlaid with either lysosomal markers or HB3-RFP fluorescence (not shown), indicating lysosome-specific localization of the Oregon Green 488-dextran. Fluorescence was visualized using a 100× S Fluor oil-immersion objective (Nikon, Japan). Samples were illuminated with 8 nm bandpass excitation at the selected wavelengths (Oregon Green 488-dextran: 490 and 440 nm, mRFP1: 550 nm). Emitted light was collected at RT for 15 s via standard dichroic mirrors and filter sets for FITC and tetramethylrhodamine respectively. 490 nm and 440 nm images were background subtracted. A region of interest (ROI) was defined around each cell expressing RFP or the PfCRT-RFP construct. Total 490 nm fluorescence within the ROI was divided by the total 440 nm fluorescence within the ROI, yielding a 490/440 ratio value representing the average ratio/pH of all loaded intracellular compartments [30]. Oregon Green 488-dextran was calibrated in situ by incubation of loaded cells in MES high-K+ buffer (5 mM NaCl, 115 mM KCl, 1.2 mM MgCl2, 25 mM MES) at varying pH and incubated with 5 μM monensin and 5 μM nigericin for 15 min. 3-8 cells at each pH were analyzed to construct the Oregon Green 488-dextran 490/440 fluorescence intensity ratio to pH calibration.
Determination of lysosomal Ca2+
HB3-RFP- or Dd2-RFP-transfected cells were re-plated onto 22 × 40 mm glass coverslips. The next day, cells were washed once with PBS then loaded in the dark for 1 hr in 1 μM Fura-2-AM (Molecular Probes, OR). Cells were washed and incubated in E1 saline buffer (140 mM NaCl, 2.8 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 2 mM EGTA, 10 mM glucose, 10 mM HEPES, pH = 7.2) for at least 30 minutes before imaging. Coverslips were mounted in a small-volume recording chamber (RC-21B, Warner Instruments, CT) and perfused continuously with E1 saline. Transfected cells were identified via their mRFP1 fluorescence excited at 555 nm. Cytoplasmic [Ca2+] was measured ratiometrically via a standard Nikon Fura-2 filter set, taking image pairs at 340 and 380 nm excitation every 0.5 s. [Ca2+]i was calibrated in situ with 2. μM ionomycin (Molecular Probes, OR) followed by perfusion with E1 saline containing 0 and 2 mM Ca2+ to define Rmin and Rmax. The 340/380 ratio to concentration calibration curve was determined by the method of Grynkiewicz et al. [31]. Drugs were applied by gravity perfusion. The images were saved to disk and played back post hoc for ROI definition and ratio calculations. Glycyl-phenylalanine-2-naphthylamide (GPN) and carbachol (CCh) were obtained from Sigma.
Measurement of radiolabeled CQ uptake by HEK293 cells
HEK293 cells were transfected as above in 100 mm dishes. After 18-24 hrs, the cells were washed twice with PBS and then removed from the dish by trypsinization. Following two washes in E1 saline, transfected cells were resuspended in 1 ml E1 saline. For each assay, 80 μl cell suspension was combined with 0.5 volumes of E1 saline containing 120 nM [3H]-CQ (specific activity 25 Ci/mmol; Amersham, NJ). Reactions were set up twice in triplicate. After addition of drug to the cells, one triplicate set of tubes was kept on ice, and the other set was incubated at 37 °C. All tubes were vortexed briefly every 15 min for 1 hr. Assays were layered carefully onto 250 μl of a 1:1 mixture of AR20/AR200 silicone oil (Sigma), and terminated by centrifugation of the cell suspension at 16,000 g for 1 min in a microfuge. The tubes were rapidly frozen on dry ice. The lower portion of the tube containing the cell pellet was sliced from the tube into a scintillation vial, and incubated at RT in 150 μl of NCS II tissue solubilizer for 10 min with vigorous shaking. 10 μl glacial acetic acid and 3.5 ml BioSafe II scintillation fluid were added, and the assays left overnight before counting. Counting was performed in an LS6500 scintillation counter (Beckman, CA). Total protein concentrations for each cell suspension were determined in triplicate using a BCA assay (Pierce, IL).
RESULTS
Heterologous expression of PfCRT in HEK293 cells
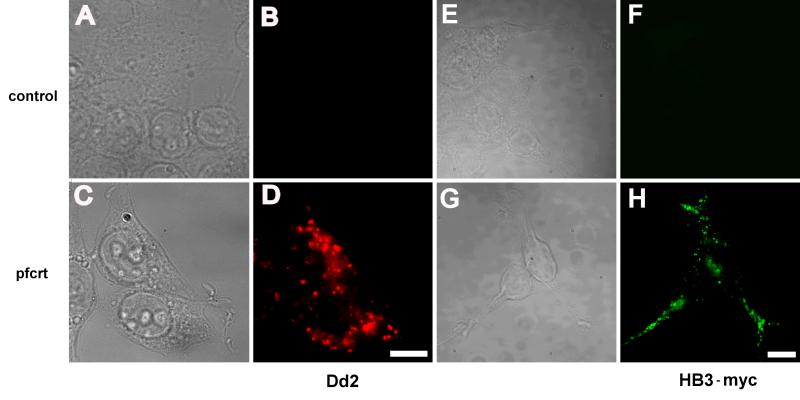
To establish a mammalian heterologous expression system to study PfCRT, we transfected HEK293 cells with yeast codon-normalized constructs expressing pfcrt alleles of the CQ-resistant Dd2 or the CQ-sensitive HB3 type [23]. These alleles differ by eight single point mutations [5]. Cells expressing PfCRT, when visualized by indirect immunofluorescence with anti-PfCRT antiserum [5], had a significant fluorescent signal (Fig. 1C, D) compared with mock-transfected cells (Fig. 1A, B). Fluorescent punctae were distributed throughout the cytoplasm, although there was some concentration in peri-nuclear areas and also in lamellipodia. There were no apparent differences between HB3 and Dd2 transfected cells (data not shown). We did not detect any fluorescence consistent with PfCRT expression on the plasma membrane in HEK293 cells (Fig. 1D, H). A small amount of diffuse non-specific background staining was seen in mock-transfected cells (Fig. 1B) and in controls where the primary antibody was omitted (data not shown). Similar punctate staining was obtained with HB3-myc (Fig. 1G, H) and Dd2-myc (data not shown) constructs.
Fig. 1.
Detection by immunofluorescence of PfCRT expression in HEK293 cells. Bright field (A, C, E, G) and epifluorescence (B, D, F, H) images of HEK293 cells, either mock transfected (A, B, E, F) or transfected with Dd2 (C, D) or HB3-myc (G, H). Cells were fixed in paraformaldehyde and permeabilized with Triton X-100. In panels B and D, mock-transfected (B) and Dd2-transfected (D) cells were probed with anti-PfCRT primary antibodies. In panels F and H, mock-transfected (F) and HB3-myc-transfected (H) cells were probed with anti-myc primary antibodies. Scale bar = 10 μm.
Subcellular localization of PfCRT in HEK293 cells
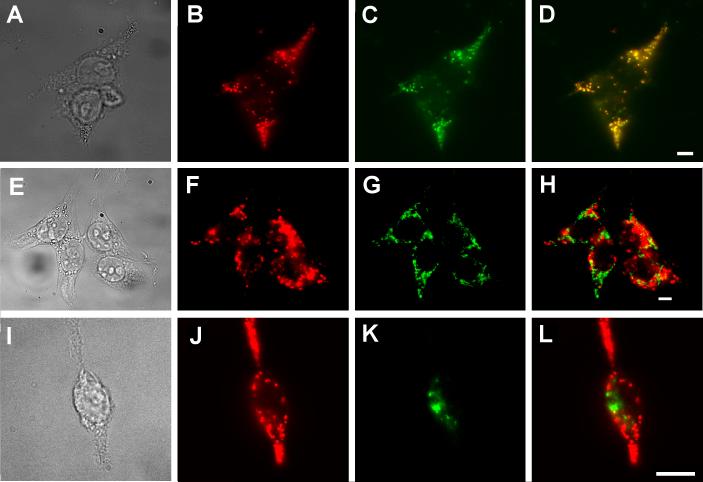
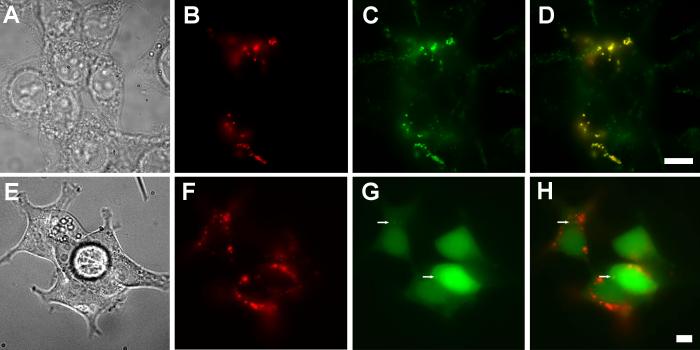
In order to determine PfCRT's subcellular localization, we performed double-labeling experiments with antibodies to organelle marker proteins and PfCRT. The fluorescence pattern in cells double-labeled with anti-PfCRT antibodies and antibodies to human lysosome-associated membrane protein 2 (LAMP-2) coincided almost exactly (Fig. 2A-D). Similarly, complete overlap was observed between the LAMP-2 marker and PfCRT fusion proteins that had either a C-terminal myc epitope tag or a monomeric Red Fluorescent Protein (mRFP1) fusion (data not shown). The peri-nuclear staining of PfCRT did not overlap with the endoplasmic reticulum marker, calnexin (Fig. 2E-H). Similarly, PfCRT did not colocalize with the late-Golgi marker GM-130 (Fig. 2I-L). We infer that PfCRT is sorted to the lysosomes in HEK293 cells.
Fig. 2.
Colocalization of PfCRT and LAMP-2. Bright field (A, E, I) and epifluorescence images (B-D, F-H, J-L) of doubly labeled, PfCRT-expressing HEK293 cells, probed with rabbit anti-PfCRT antibodies detected via Cy3-secondary anti rabbit IgG antibodies (B, F, J; red), as well as mouse monoclonal antibodies against LAMP-2 (C), calnexin (G) or GM-130 (K), detected with Alexa Fluor 488-labeled secondary anti-mouse IgG antibodies (green). (D, H, L) Overlays of the red and green channels. Yellow indicates areas of overlap. Scale bar = 10 μm.
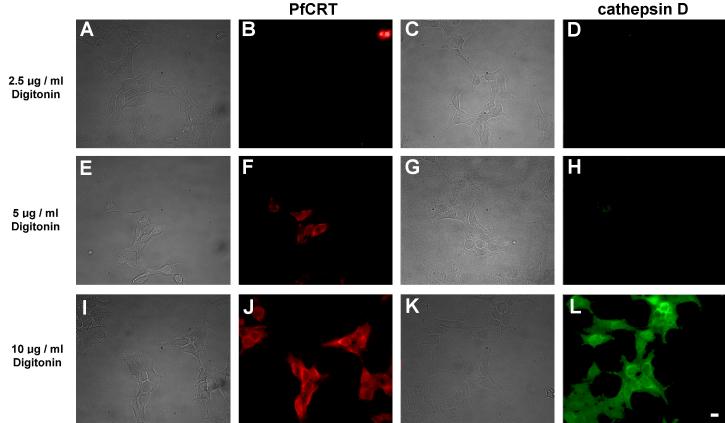
For PfCRT to be functional it should be in the limiting lysosomal membrane and not inside the lysosomes where it may be destined for sequestration or degradation, for example by incorporation in multivesicular bodies within the lysosome [32]. The optical resolution of immunofluorescence is insufficient to distinguish between localization in the membrane vs. inside the lysosome. Therefore, we sought to demonstrate that PfCRT's C-terminus was exposed in the cytoplasm, which would be consistent with localization in the lysosomal membrane. Digitonin was used to selectively permeabilize the plasma membrane without disturbing the integrity of the lysosomal membrane [29]. We demonstrated selective plasma membrane permeabilization using tubulin as a cytoplasmic marker and cathepsin as an intra-lysosomal marker. Treating cells with 5 μg/ml digitonin permeabilized the plasma membrane but not the lysosomal membrane. Consistent with this, cells treated with 5 μg/ml digitonin were labeled by anti-tubulin (data not shown) but not by anti-cathepsin antibodies (Fig. 3H), implying that only the plasma membrane was permeabilized. In contrast, 10 μg/ml digitonin or Triton X-100 permeabilize both the plasma membrane and the lysosomal membrane. Cells treated with 10 μg/ml digitonin were labeled with both anti-tubulin (data not shown) and anti-cathepsin (Fig. 3L) antibodies, implying permeabilization of both membranes. In order to determine whether PfCRT was present in the lysosomal membrane we used Dd2-bad, a construct that expresses PfCRT with the biotin-acceptor domain fused to its C-terminus, which is predicted to be cytoplasmic. Given the predicted cytoplasmic location, the biotin-acceptor domain should be biotinylated and thus detectable with a fluorescent streptavidin derivative [23]. Following permeabilization of just the plasma membrane with 5 μg/ml digitonin, Dd2-bad-expressing cells were labeled with Cy3-streptavidin (Fig. 3F). The presence of punctate Cy3 fluorescence in these plasma membrane permeabilized cells indicated that the biotinylated C-terminus of PfCRT was exposed on the cytoplasmic side of the lysosomal membrane. Therefore, we inferred that PfCRT was located in the lysosomal limiting membrane with its C-terminus in the cytoplasm. This implies that the lysosomal localization of PfCRT in HEK293 cells is due to specific targeting to this subcellular membrane compartment.
Fig. 3.
Demonstration of selective plasma membrane permeabilization by digitonin. HEK293 cells transfected with Dd2-bad were fixed and permeabilized with digitonin at the concentration indicated on the left of each row. Bright field images are shown on the left hand side of each corresponding epifluorescence image. (B, F, J) Red fluorescence from Cy3-streptavidin. (D, H, L) Green fluorescence from cathepsin D labeled with anti-cathepsin primary antibodies and Alexa Fluor 488 secondary antibodies. For cells treated with 5 μg/ml digitonin, note the red fluorescence in panel F indicating detection of the biotinylated PfCRT C-terminal fusion biotin-acceptor domain, and the corresponding lack of green fluorescence in panel H at the same digitonin concentration indicating the inaccessibility of intralysosomal cathepsin D to the anti-cathepsin antibodies. This indicates that the plasma membrane has been permeabilized but the lysosomal membrane has not been permeabilized. The ability of Cy3-streptavidin to bind to the C-terminal fusion biotin-acceptor domain under these conditions indicates that PfCRT is embedded in the limiting membrane of the lysosomes with its C-terminus in the cytoplasm. At 10 μg/ml digitonin the lysosomal membrane was permeabilized and the anti-cathepsin antibodies show extensive labeling (L). Scale bar = 10 μm.
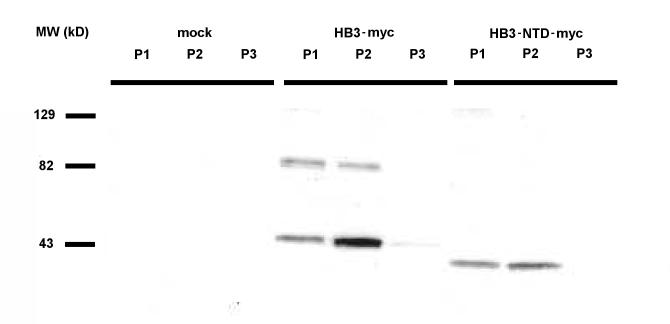
Subcellular fractionation by differential centrifugation supported PfCRT's lysosomal localization. A low-speed spin gave a nuclear pellet (P1), a medium-speed spin pelleted lysosomes and mitochondria (P2), and a high-speed spin pelleted the remaining membranes (P3). Cells transfected with HB3-myc were fractionated in this way, and Western blots of subcellular fractions were visualized with an anti-myc monoclonal antibody (Fig. 4). No cross-reactions were observed in lysates from mock-transfected cells. For transfected cells, we observed a strong band in the P2 fraction at approximately 46 kDa, and much weaker bands at the same Mr in the P1 and P3 fractions. Our estimate of 46 kDa for the Mr of HB3-myc was consistent with a predicted molecular weight of 52 kDa, as integral membrane proteins often migrate anomalously in SDS-PAGE gels. We assume that the weaker bands represent contamination with a small proportion of lysosomal material, as we did not detect PfCRT in the plasma membrane or nucleus by immunofluorescence (Fig. 1). We also observed weak bands in the P1 and P2 fractions at approximately 90 kDa. These bands likely represent aggregation of the protein, as heating of the samples above RT or boiling before electrophoresis prevented HB3-myc protein from entering the gel (data not shown).
Fig. 4.
Subcellular fractionation of HEK293 cells expressing PfCRT. Western blot of transfected HEK293 cell fractions. Cells were mock-transfected (water), or transfected with HB3-myc or HB3-NTD-myc. The blot was probed with anti-myc tag antibodies, and developed via ECL. Key to fractions: P1 = 800 g pellet, P2 = 14,000 g pellet, P3 = 100,000 g pellet. Note that, as expected, the molecular weight of the N-terminal domain deletion (HB3-NTDmyc) construct is about 5 kDa less than full length PfCRT (HB3-myc).
Lysosomes are a site of protein degradation. To confirm that PfCRT was not being digested by HEK293 cell lysosomes, we fractionated cells expressing HB3-NTD-myc, a construct that deletes approximately 5 kDa from the N-terminus of PfCRT (Fig. 4). This construct was detected via its C-terminal myc tag and had an apparent Mr of approximately 41 kDa. This is about 5 kDa smaller than full length PfCRT. Therefore, we infer that PfCRT was not digested in the lysosomes, as C-terminal tags remained attached (as shown by detection through the myc tag) and the N-terminal deletion mutant HB3-NTD-myc migrated at an Mr approximately 5 kDa less than HB3-myc (Fig. 4), consistent with the N-terminal sequence being retained in the full length HB3-myc homogenates.
PfCRT expression caused acidification of the pH in lysosomes
Previous studies have indicated a potential effect of PfCRT expression on vesicular pH in heterologous expression systems [18, 23]. Furthermore, CQR parasites may have a more acidic DV pH [20]; this finding however has been debated [11, 19, 20, 22, 33]. Therefore, we measured the lysosomal pH in individual cells transfected with HB3-RFP and Dd2-RFP, compared with control cells transfected with RFP alone. C-terminally RFP-tagged protein was used to identify PfCRT expressing cells at the beginning of live-cell imaging experiments, without significant fluorescence emission overlap with the probes used for measurement of lysosomal pH and [Ca2+]i. RFP-tagged PfCRT constructs localized to LAMP-2-positive vesicles in a manner similar to untagged proteins (data not shown) and was co-localized with fluorescent Oregon Green 488 dextran. We infer that the C-terminal RFP fusion protein was sorted to lysosomes just as the wild type and myc-tagged constructs.
Lysosomal pH in HEK293 cells was measured by live cell ratiometric imaging of Oregon Green 488-dextran, which accumulates in lysosomes following endocytosis from the bath solution and has a pKa of approximately 4.7 (Fig. 5A) (http://probes.invitrogen.com/servlets/datatable?item=6146&id=28860). The mean pH measured in 5 to 15 cells varied slightly between transfections. The variance of the mean (S.D.) of five different transfections was 0.25 ± 0.01 pH units (n=5). The mean pH values for the RFP-transfected control cells were variable between transfections, but the variance within each transfection did not significantly differ from the mean variance for all transfections (Bartlett's test for equal variances, p > 0.05). The average intralysosomal pH in the RFP-transfected control cells for five transfections (69 cells) was 4.80 ± 0.03. We observed a consistent significant acidification of 0.26 pH units in lysosomes in cells expressing HB3-RFP (4.54 ± 0.03) compared with those expressing RFP in the absence of PfCRT (Fig. 5B). The lysosomal pH was even more acidified in cells expressing Dd2-RFP (4.30 ± 0.03) where the pH was 0.50 pH units lower than in the RFP transfected control cells (Fig. 5B). The lysosomes in Dd2-RFP-expressing cells were significantly more acidic (0.24 pH units) than those in HB3-RFP-expressing cells (Fig. 5B, one way ANOVA with Bonferroni's Multiple Comparison post-test, p < 0.001, n ≥ 15).
Fig. 5.
Measurement of lysosomal pH in cells transfected with pfcrt. (A) Calibration curve for the pH dependent fluorescence of Oregon Green 488 dextran in lysosomes. Cells loaded with Oregon Green 488 dextran were equilibrated with solutions of known pH in the presence of the ionophores nigericin and monensin. The ratio of the fluorescence emission intensity of the Oregon Green 488 dextran loaded cells due to excitation at 490 and 440 nm was determined as a function of bath solution pH. (B) The lysosomal pH is more acidic in cells expressing the PfCRT isoforms HB3-RFP or Dd2-RFP than in RFP transfected control cells. Lysosomal pH was measured in live cells using Oregon Green 488 dextran. The RFP fluorescence allowed rapid identification of transfected cells. The mean lysosomal pH for HB3-RFP- or Dd2-RFP-transfected cells are shown, along with mean pH measured for the reversal mutants HB3-RFP-K76T and Dd2-RFP-T76K. Control cells were transfected with RFP alone. Error bars indicate S.E.M. and asterisks (*) indicate significant differences from control, as determined by one way ANOVA with Bonferroni multiple comparison post-test (p < 0.05).
Numerous studies have implicated the PfCRT residue at position 76 as being key to CQ resistance [3, 10]. CQR patient isolates have a threonine at this position whereas CQS isolates have a lysine. We sought to determine whether mutation at this position altered the extent of lysosomal acidification. Cells expressing HB3-RFP-K76T had a lysosomal pH of 4.26 ± 0.03 (Fig 5B). This represents a significant acidification of HB3-RFP-K76T lysosomal pH by 0.54 pH units compared to control cells. This lysosomal acidification was similar to that measured in Dd2-RFP-transfected cells. Thus, in the HB3 background, the K76T mutation was sufficient to confer the increased acidification observed in Dd2 expressing cells.
Conversely, in cells expressing the Dd2 back mutant, Dd2-RFP-T76K, the lysosomal pH was 4.67 ± 0.05 (Fig 5B). This was significantly less acidic than that observed in cells expressing Dd2. This pH was not significantly different from the pH in cells transfected with HB3-RFP or with RFP alone. Thus, the amino acid at position 76 is a critical determinant of the increased acidification that we observed in Dd2 expressing cells.
We infer that PfCRT expression results in acidification of the lysosomes in HEK293 cells, and that a pfcrt allele that causes CQ resistance in parasites, Dd2, causes greater lysosomal acidification than the CQS allele, HB3. Furthermore, reversal mutations in these alleles at residue 76 that were previously shown to be critical to CQ resistance in Plasmodium itself [10], mirror their functional effects on lysosomal pH in this heterologous expression system.
PfCRT expression did not affect lysosomal Ca2+ release
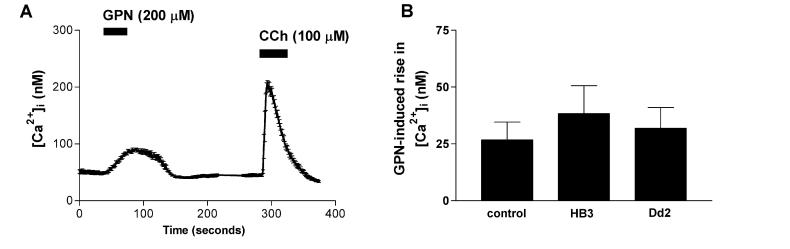
To test whether the effects of PfCRT expression on lysosomal pH were specific or part of more general changes in lysosomal function we assayed the steady state lysosomal Ca2+ content. Ca2+ ions have also been suggested as a potential modifier of parasite drug resistance in the P. falciparum DV [34]. Therefore, we measured the steady-state Ca2+ content of lysosomes in HEK293 cells expressing PfCRT isoforms (Fig. 6). We detected the release of lysosomal Ca2+ into the cytoplasm of cells transfected with HB3-RFP or Dd2-RFP by Fura-2 imaging of cytoplasmic [Ca2+]. The application of GPN caused the reversible and specific osmotic swelling of lysosomes, releasing small molecules including Ca2+ into the cytoplasm [35] (Fig. 6A). Subsequent carbachol application after GPN washout demonstrated the release of Ca2+ from conventional endoplasmic reticulum stores (Fig. 6A). There was no significant difference in the background-subtracted peak GPN-induced lysosomal Ca2+ release (27 – 38 nM) from cells transfected with either pfcrt allele, or from mock transfected cells (Fig. 6B, one way ANOVA, p > 0.05, n ≥ 6), suggesting that PfCRT expression did not significantly alter lysosomal [Ca2+].
Fig. 6.
GPN-induced lysosomal Ca2+ release monitored by Fura-2 imaging. Cells were loaded with Fura-2-AM and the cytoplasmic Ca2+ concentration was determined continuously by ratiometric imaging. (A) Mean cytoplasmic Ca2+ of seven HEK293 cells in a field during application of GPN (200 μM) and carbachol (100 μM). (B) Mean peak lysosomal Ca2+ release by GPN from HEK293 cells transfected with RFP controls, HB3-RFP or Dd2-RFP. The means are not significantly different. Error bars indicate S.E.M.
PfCRT expression did not affect CQ accumulation
Several studies have shown that CQ accumulation is reduced in CQR P. falciparum strains. Therefore, we measured the accumulation of [3H]-CQ into HEK293 cells transfected with either pcDNA3.1(+) empty vector (control) or HB3 or Dd2 constructs. With an external [CQ] of 40 nM, control cells accumulated 13.2 ± 1.6 fmol/μg protein (mean from 3 independent transfections). HB3-transfected cells accumulated 12.6 ± 2.4 fmol/μg protein, while Dd2-transfected cells accumulated 11.4 ± 1.6 fmol/μg protein. There was no significant difference between either HB3 or Dd2 and control transfected cells (one way ANOVA with Bonferroni post-test, p > 0.05). These experiments were performed using transiently transfected populations of cells, and we routinely observed transfection efficiencies of greater than 85% in our hands, therefore we believe these data to reasonably reflect the mean effect on PfCRT-expressing HEK293 cells.
Evaluation of lysosomal sorting signals in PfCRT
We sought to identify the sequence or structural motif(s) within PfCRT that resulted in targeting to the lysosomal membrane. Many of the known mechanisms for membrane protein sorting to lysosomes rely on signal recognition by cytosolic proteins [36-38]. Thus, the sorting signals might be expected to reside in PfCRTfs putative cytoplasmic domains. These are predicted to include the N- and C-termini and four cytoplasmic loops. Lysosomal targeting signals include dileucine, NXPY and YXXΦ motifs, and ubiquitylation. There are no dileucine or NXPY motifs in the putative cytoplasmic loops, however, there is a YXXΦ motif near the N terminus (Y20-L23). Using deletion constructs we tested whether lysosomal targeting sequences resided in the N- or C-terminal domains. Removal of the majority of either or both of the predicted N- or C-terminal cytoplasmic domains did not substantially alter PfCRT localization (Fig. 7A-D).
Fig. 7.
Attempted perturbation of PfCRT sorting. Truncation of the N- and C-terminal domains does not alter PfCRT trafficking to lysosomes nor does coexpression of PfCRT with LC3-GFP. (A-D) HB3-NTD-CTD-myc PfCRT that lacks the N- and C-terminal cytoplasmic domains still traffics to the LAMP-2 positive lysosomal compartment. (A) Bright field, (B; red) HB3-NTD-CTD-myc, detected via indirect immunofluorescence with an anti-myc-tag antibody, biotinylated-anti-rabbit secondary and Cy3-streptavidin. (C; green) LAMP-2, detected via H4B4 antibodies and anti-mouse Alexa Fluor 488 secondary antibodies. (D) Overlay of panels B and C showing colocalization of LAMP-2 and HB3-NTD-CTD-myc. (E-H) Illustrates that overexpression of LC3, which causes an increase in autophagic vesicle formation, does not alter PfCRT localization. (E) Bright field, (F, red) detected via indirect immunofluorescence with anti-myc-tag antibodies, biotinylated-anti-rabbit secondary and Cy3-streptavidin; (G; green) Green fluorescence from LC3-GFP. (H) Overlay of panels F and G. Arrows indicate punctae of LC3-GFP. Scale bar = 10 μm.
Transient mono- to tri-ubiquitylation can be sufficient for lysosomal targeting [36]. Neither the 45 kDa band detected in Plasmodium [5] nor the 46 kDa protein that we have detected following PfCRT expression in HEK293 cells appear to have the requisite higher Mr that would indicate ubiquitylation, compared with the predicted Mr of 52 kDa. Nevertheless, because the lysosomal targeting can arise from transient ubiquitylation we mutated all putative cytoplasmic loop lysine residues to arginine (HB3-kminus) to prevent ubiquitylation. This mutant remained co-localized with LAMP-2 (data not shown).
As PfCRT sorting was not affected by deletion of known lysosomal targeting sequences, we attempted to subvert non-sequenced-based sorting by co-expression of PfCRT with dominant negative mutants of proteins implicated in lysosomal sorting. Co-expression of a dominant negative tetraspanin mutant, CD63-AEVM [39], did not alter PfCRT targeting to the lysosome (not shown). We noted that the P. falciparum genome sequence [40] contains homologues of human vacuolar protein sorting protein VPS4 [41, 42] and human sorting nexin 4 (SNX4) [43]. Co-expression of dominant negative mutants of either of these proteins did not perturb PfCRT sorting to the lysosome. LC3-GFP overexpression is known to cause the formation of autophagic vesicles [44] but when co-expressed with PfCRT, the latter did not co-localize with the punctate structures induced by LC3-GFP expression (Fig. 7E-H). Thus, at present we are unable to identify the lysosomal targeting motifs within PfCRT. One possible explanation is that the predicted transmembrane topology may be incorrect and one of the six YXXΦ motifs predicted to be in luminal loops or transmembrane segments or the dileucine near the luminal end of TM10 is actually in a cytoplasmic loop. Experimental verification of the predicted transmembrane topology may help to resolve this issue.
DISCUSSION
We report the successful expression of PfCRT protein in an experimentally accessible mammalian cell system with a view to characterizing its structure and function. While the discovery [5] and subsequent study [see for review 11] of the pfcrt gene has added much to our understanding of the molecular mechanism of CQ resistance in malaria, neither the endogenous function, nor the physiological mechanism by which pfcrt confers CQ resistance are clear. Heterologous expression of PfCRT in Pichia pastoris, Saccharomyces cerevisiae [23, 24], Dictyostelium discoides [18] and Xenopus laevis [25] has shown that the protein is more amenable to biochemical and physiological analysis in less technically onerous systems than P. falciparum. We chose the HEK293 cell line for heterologous expression, as it is widely used for functional studies of cellular physiology. This is the first report of PfCRT heterologous expression in a mammalian cell line.
In HEK293 cells, heterologously expressed PfCRT was almost entirely located in the lysosomes based on colocalization with the lysosomal marker LAMP-2 and subcellular fractionation. PfCRT was embedded in the lysosomal limiting membrane with its C-terminus exposed to the cytoplasm as indicated by selective permeabilization experiments and by biotinylation of a biotin-acceptor-domain fused to the C-terminus. Therefore, PfCRT was not inside the lysosomes destined for degradation. The lysosomes are the analogous intracellular organelle to the parasite's DV where PfCRT is found in situ [5]. Both the lysosomes and DV are acidic, proteolytic compartments. Thus, in the HEK293 heterologous expression system PfCRT is localized in a membrane environment similar to its native environment in the P. falciparum DV, where it is exposed to a large transmembrane proton gradient.
Using a single cell ratiometric fluorescence assay in which lysosomes were loaded with Oregon Green-dextran, we showed that expression of the CQR Dd2 isoform resulted in significantly greater lysosomal acidification than expression of the CQS HB3 isoform. The amino acid residue at position 76 is directly correlated with CQ resistance in the parasite itself [3, 10]. In CQS patient isolates a lysine residue is present at this position, whereas, in CQR patient isolates the lysine is replaced by threonine [11]. Furthermore, in a CQR line developed in the lab under drug pressure, an isoleucine was found at position 76 [5, 45]. The point mutation K76T in the CQS HB3 background caused a similar extent of lysosomal acidification as Dd2. Conversely, the T76K mutation in the Dd2 background resulted in significantly less acidification than Dd2. Thus, the extent of lysosomal acidification was correlated with the amino acid at position 76 regardless of the HB3 or Dd2 background. Therefore, the extent of lysosomal acidification provided a direct functional assay for the effects of mutations in PfCRT. In P. falciparum the Dd2-T76K mutant is CQS, whereas attempts to insert the HB3-K76T mutant into parasites by allelic exchange did not lead to viable parasites; the basis for this failure is unknown [10].
In intact parasites, the DV pH has been reported to be 0.4-0.7 pH units lower in CQR compared with CQS lines [20, 46]. A separate study however, reported a slight alkalinization of CQR parasite DVs compared with CQS parasites, although this difference disappeared at lower pH-sensitive dye concentrations [33]. These discrepancies in reported DV pH may be more reflective of differences in measurement techniques than of physiological variations in parasites. Notwithstanding, several previous reports on the heterologous expression of pfcrt have noted that PfCRT expression causes acidification of either inside-out plasma membrane vesicles [23] or acidic vacuoles [0.1-0.2 pH units, 18]. Our results are consistent with these previous findings and extend them by showing that the extent of acidification is directly related to the presence or absence of a lysine at position 76. These data provide a strong association between the lysosomal acidification phenomenon observed in our heterologous expression system with CQ sensitivity data obtained from P. falciparum containing similar mutations [10]. Interestingly, FPIX solubility decreases at more acidic pH [47], therefore vacuolar acidification may contribute to the CQ resistance phenotype in the parasite but this remains to be proven. Furthermore, our finding that the presence of the pfcrt HB3 (CQS) allele leads to lysosomal acidification is novel, and suggests that whatever the process by which PfCRT causes organelle acidification, it operates in both CQS and CQR forms of this transmembrane protein, albeit to a different extent. The lysosomal acidification that we observed in human cells could be due to a component of the endogenous function of PfCRT in the parasite DV, in addition to being a correlate of CQ resistance.
How could PfCRT expression cause increased acidification of the intralysosomal space in HEK293 cells? In order to enumerate the possible explanations one must first examine the functional elements in the acidification process. In vivo lysosomal acidification is an ATP-dependent process that runs down quickly in the absence of ATP. To maintain the steady state acidic pH involves the activities of at least three types of proteins: 1) an ATP-coupled H+ transporter (V-type H+-ATPase), 2) an H+ leak pathway (to explain the pH gradient rundown in the absence of ATP or following inhibition of the H+-ATPase), and 3) a non-H+ permeable electrical shunt conductance (the electrogenic V-Type H+-ATPase will create a membrane potential, Δψ, across the lysosomal membrane; this membrane potential must be shunted to allow sufficient accumulation of protons to create a proton concentration gradient across the lysosomal membrane) [48, 49]. The lysosomal pH is maintained in a steady state. It is not known to what extent each of these three processes controls the steady state pH level [49]. PfCRT expression could cause lysosomal acidification by 1) modulating the activity of an endogenous lysosomal protein involved in lysosomal acidification, 2) forming a non-H+ permeable shunt conductance or 3) functioning as an energy-coupled H+ transporter, in which case H+ transport would need to be coupled to an energy source or ion gradient to drive the uphill movement of protons from the cytoplasm into the lysosomes to cause acidification. At present we cannot distinguish between these possibilities, but further studies of isolated lysosomes containing PfCRT may allow us to distinguish these alternatives. Bioinformatic studies have linked PfCRT to a variety of transporters but have not provided any clear indication of PfCRT's endogenous substrates [7]. Thus protons remain a candidate substrate. While the identity of the endogenous substrate or substrates for PfCRT remains elusive, our model system and assay presented here will be useful in screening among candidates.
The changes in intralysosomal pH associated with PfCRT expression were not due to a generalized change in lysosomal ion transport, as we detected no significant difference between the peak lysosomal Ca2+ release induced by GPN in mock-transfected cells compared with either HB3-RFP- or Dd2-RFP-transfected cells, an observation consistent with that obtained for the parasite DV [50].
Does PfCRT mediate chloroquine efflux in HEK293 cells?
In contrast to the striking effects of PfCRT on lysosomal pH in HEK293 cells, we found no significant difference between the levels of [3H]-CQ accumulation in cells transfected with PfCRT constructs. This result is similar to that found for inside-out plasma membrane vesicles from yeast cells expressing PfCRT [24]. Our data in HEK293 cells does not provide evidence for a role of PfCRT in CQ efflux from lysosomes. Our finding contrasts with a recent study performed under similar conditions that demonstrated that heterologous expression of PfCRT constructs in Dictyostelium conferred a CQ efflux phenotype [18]. We cannot currently explain this discrepancy, but we speculate that in contrast to human cells, Dictyostelium itself expresses PfCRT homologues, it may therefore contain as yet unidentified biochemical component(s) necessary for the CQ efflux phenotype, which may not be present in HEK293 cells. Alternatively, PfCRT may not directly transport CQ. Our finding that PfCRT alone is not sufficient for CQ efflux suggests that the mechanism of CQ efflux in CQR P. falciparum may be more complicated than previously envisaged.
PfCRT trafficking to lysosomes
The absence of PfCRT in the plasma membrane was surprising because this is the default location for trafficking of integral membrane proteins, and two previous studies have reported plasma membrane expression of heterologously expressed PfCRT [23, 25]. In Xenopus laevis oocytes, the close proximity of the endoplasmic reticulum and Golgi apparatus to the plasma membrane makes it difficult to definitively establish plasma membrane localization using immunofluorescence microscopy on frozen sections [51]. We cannot rule out the possibility that in HEK293 cells PfCRT may be transiently exported to the plasma membrane and then rapidly internalized during trafficking to the lysosomes. If this were occurring, then the amount of PfCRT in the plasma membrane is below our level of detection.
In Dictyostelium, PfCRT “mislocalized throughout the cell” [18]; and only a chimera formed by substituting the N-terminal domain from a homologous Dictyostelium protein for the N-terminus of PfCRT resulted in localization of this protein to acidic vesicles. The role of the N-terminal domain in PfCRT trafficking may depend on the heterologous expression system because in HEK293 cells deletion of the N-terminus had no effect on PfCRT trafficking.
The successful heterologous expression of functional, epitope-tagged PfCRT in HEK293 cells presented here represents a novel system to facilitate the study of molecular structure and function of both in situ and purified PfCRT. This will greatly facilitate investigations of the CQ resistance mechanism and may provide a basis for assay development to identify new drugs to combat the growing global threat of drug-resistant malaria.
ACKNOWLEDGEMENTS
We thank Moez Bali, Amal Bera, Ana Maria Cuervo, Alan Finkelstein, Jeffrey Horenstein, Michaela Jansen, David Johnson, Sean McBride, Nicole McKinnon, Paul Riegelhaupt and Dennis Shields for helpful discussions. We thank Drs. David Sharp and Philip Aisen for use of cell culture facilities, Dr. Dennis Shields for GM-130 antibody, and Dr. Roger Tsien for mRFP1 cDNA.
Abbreviations
- bad
biotin acceptor domain
- [Ca]i
intracellular calcium concentration
- CQ
chloroquine
- CQR
chloroquine resistant
- CQS
chloroquine sensitive
- DV
digestive vacuole
- FPIX
ferriprotoporphyrin IX
- GPN
Glycyl-phenylalanine-2-naphthylamide
- HEK293
human embryonic kidney cell
- LAMP
Lysosome-associated membrane protein
- PfCRT
Plasmodium falciparum chloroquine resistance transporter
- RFP
monomeric red fluorescent protein, mRFP1
- ROI
region of interest
- RT
room temperature
Footnotes
This work was supported in part by NIH grants AI952710 (M.H.A.), AI50234 (D.A.F.), AI045957 (P.D.R.) and AI056312 (P.D.R.) and an Award from the Speaker's Fund for Biomedical Research, New York Academy of Medicine (D.A.F.). D.A.L. was supported in part by the Medical Scientist Training Program Grant for the NIH (GM007288).
REFERENCES
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talisuna AO, Bloland P, D'Alessandro U. History, dynamics, and public health importance of malaria parasite resistance. Clin Microbiol Rev. 2004;17:235–254. doi: 10.1128/CMR.17.1.235-254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 4.Olliaro PL, Taylor WRJ. Antimalarial compounds: from bench to bedside. J Exp Biol. 2003;206:3753–3759. doi: 10.1242/jeb.00653. [DOI] [PubMed] [Google Scholar]
- 5.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LMB, Sidhu ABS, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. Mutations in the P-falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidhu ABS, Verdier-Pinard D, Fidock DA. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science. 2002;298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin RE, Kirk K. The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol Biol Evol. 2004;21:1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- 8.Tran CV, Saier MH., Jr The principal chloroquine resistance protein of Plasmodium falciparum is a member of the drug/metabolite transporter superfamily. Microbiology. 2004;150:1–3. doi: 10.1099/mic.0.26818-0. [DOI] [PubMed] [Google Scholar]
- 9.Johnson DJ, Fidock DA, Mungthin M, Lakshmanan V, Sidhu AB, Bray PG, Ward SA. Evidence for a central role for PfCRT in conferring Plasmodium falciparum resistance to diverse antimalarial agents. Mol Cell. 2004;15:867–877. doi: 10.1016/j.molcel.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakshmanan V, Bray PG, Verdier-Pinard D, Johnson DJ, Horrocks P, Muhle RA, Alakpa GE, Hughes RH, Ward SA, Krogstad DJ, Sidhu AB, Fidock DA. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray PG, Martin RE, Tilley L, Ward SA, Kirk K, Fidock DA. Defining the role of PfCRT in Plasmodium falciparum chloroquine resistance. Mol Microbiol. 2005;56:323–333. doi: 10.1111/j.1365-2958.2005.04556.x. [DOI] [PubMed] [Google Scholar]
- 12.Fitch CD. Ferriprotoporphyrin IX, phospholipids, and the antimalarial actions of quinoline drugs. Life Sci. 2004;74:1957–1972. doi: 10.1016/j.lfs.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Saliba KJ, Folb PI, Smith PJ. Role for the Plasmodium falciparum digestive vacuole in chloroquine resistance. Biochem Pharmacol. 1998;56:313–320. doi: 10.1016/s0006-2952(98)00140-3. [DOI] [PubMed] [Google Scholar]
- 14.Bray PG, Mungthin M, Ridley RG, Ward SA. Access to hematin: the basis of chloroquine resistance. Mol Pharmacol. 1998;54:170–179. doi: 10.1124/mol.54.1.170. [DOI] [PubMed] [Google Scholar]
- 15.Bray PG, Janneh O, Raynes KJ, Mungthin M, Ginsburg H, Ward SA. Cellular uptake of chloroquine is dependent on binding to ferriprotoporphyrin IX and is independent of NHE activity in Plasmodium falciparum. J Cell Biol. 1999;145:363–376. doi: 10.1083/jcb.145.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez CP, McLean JE, Rohrbach P, Fidock DA, Stein WD, Lanzer M. Evidence for a pfcrt-associated chloroquine efflux system in the human malarial parasite Plasmodium falciparum. Biochemistry. 2005;44:9862–9870. doi: 10.1021/bi050061f. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez CP, Stein W, Lanzer M. Trans stimulation provides evidence for a drug efflux carrier as the mechanism of chloroquine resistance in Plasmodium falciparum. Biochemistry. 2003;42:9383–9394. doi: 10.1021/bi034269h. [DOI] [PubMed] [Google Scholar]
- 18.Naude B, Brzostowski JA, Kimmel AR, Wellems TE. Dictyostelium discoideum expresses a malaria chloroquine resistance mechanism upon transfection with mutant, but not wild-type, Plasmodium falciparum transporter PfCRT. J Biol Chem. 2005;280:25596–25603. doi: 10.1074/jbc.M503227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spiller DG, Bray PG, Hughes RH, Ward SA, White MR. The pH of the Plasmodium falciparum digestive vacuole: holy grail or dead-end trail? Trends Parasitol. 2002;18:441–444. doi: 10.1016/s1471-4922(02)02365-6. [DOI] [PubMed] [Google Scholar]
- 20.Bennett TN, Kosar AD, Ursos LMB, Dzekunov S, Sidhu ABS, Fidock DA, Roepe PD. Drug resistance-associated pfCRT mutations confer decreased Plasmodium falciparum digestive vacuolar pH. Mol Biochem Parasitol. 2004;133:99–114. doi: 10.1016/j.molbiopara.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Martin SK, Oduola AM, Milhous WK. Reversal of chloroquine resistance in Plasmodium falciparum by verapamil. Science. 1987;235:899–901. doi: 10.1126/science.3544220. [DOI] [PubMed] [Google Scholar]
- 22.Wissing F, Sanchez CP, Rohrbach P, Ricken S, Lanzer M. Illumination of the malaria parasite plasmodium falciparum alters intracellular pH - Implications for live cell imaging. J Biol Chem. 2002;277:37747–37755. doi: 10.1074/jbc.M204845200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H, Howard EM, Roepe PD. Analysis of the antimalarial drug resistance protein Pfcrt expressed in yeast. J Biol Chem. 2002;277:49767–49775. doi: 10.1074/jbc.M204005200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Paguio M, Roepe PD. The antimalarial drug resistance protein Plasmodium falciparum chloroquine resistance transporter binds chloroquine. Biochemistry. 2004;43:8290–8296. doi: 10.1021/bi049137i. [DOI] [PubMed] [Google Scholar]
- 25.Nessler S, Friedrich O, Bakouh N, Fink RH, Sanchez CP, Planelles G, Lanzer M. Evidence for activation of endogenous transporters in Xenopus laevis oocytes expressing the Plasmodium falciparum chloroquine resistance transporter, PfCRT. J Biol Chem. 2004;279:39438–39446. doi: 10.1074/jbc.M404671200. [DOI] [PubMed] [Google Scholar]
- 26.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen CA, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 29.Kyttala A, Ihrke G, Vesa J, Schell MJ, Luzio JP. Two motifs target Batten disease protein CLN3 to lysosomes in transfected nonneuronal and neuronal cells. Mol Biol Cell. 2004;15:1313–1323. doi: 10.1091/mbc.E03-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vergne I, Constant P, Laneelle G. Phagosomal pH determination by dual fluorescence flow cytometry. Anal Biochem. 1998;255:127–132. doi: 10.1006/abio.1997.2466. [DOI] [PubMed] [Google Scholar]
- 31.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 32.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 33.Hayward R, Saliba KJ, Kirk K. The pH of the digestive vacuole of Plasmodium falciparum is not associated with chloroquine resistance. J Cell Sci. 2006;119:1016–1025. doi: 10.1242/jcs.02795. [DOI] [PubMed] [Google Scholar]
- 34.Biagini GA, Bray PG, Spiller DG, White MRH, Ward SA. The digestive food vacuole of the malaria parasite is a dynamic intracellular Ca2+ store. J Biol Chem. 2003;278:27910–27915. doi: 10.1074/jbc.M304193200. [DOI] [PubMed] [Google Scholar]
- 35.Haller T, Dietl P, Deetjen P, Volkl H. The lysosomal compartment as intracellular calcium store in MDCK cells: a possible involvement in InsP3-mediated Ca2+ release. Cell Calcium. 1996;19:157–165. doi: 10.1016/s0143-4160(96)90084-6. [DOI] [PubMed] [Google Scholar]
- 36.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 37.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 38.Luzio JP, Poupon V, Lindsay MR, Mullock BM, Piper RC, Pryor PR. Membrane dynamics and the biogenesis of lysosomes. Mol Membr Biol. 2003;20:141–154. doi: 10.1080/0968768031000089546. [DOI] [PubMed] [Google Scholar]
- 39.Duffield A, Kamsteeg EJ, Brown AN, Pagel P, Caplan MJ. The tetraspanin CD63 enhances the internalization of the H,K-ATPase beta-subunit. Proc Natl Acad Sci USA. 2003;100:15560–15565. doi: 10.1073/pnas.2536699100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DMA, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang M, Coppens I, Wormsley S, Baevova P, Hoppe HC, Joiner KA. The Plasmodium falciparum Vps4 homolog mediates multivesicular body formation. J Cell Sci. 2004;117:3831–3838. doi: 10.1242/jcs.01237. [DOI] [PubMed] [Google Scholar]
- 42.Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol Biol Cell. 2000;11:227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002;3:919–931. doi: 10.1038/nrm974. [DOI] [PubMed] [Google Scholar]
- 44.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cooper RA, Ferdig MT, Su XZ, Ursos LM, Mu J, Nomura T, Fujioka H, Fidock DA, Roepe PD, Wellems TE. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- 46.Dzekunov SM, Ursos LM, Roepe PD. Digestive vacuolar pH of intact intraerythrocytic P. falciparum either sensitive or resistant to chloroquine. Mol Biochem Parasitol. 2000;110:107–124. doi: 10.1016/s0166-6851(00)00261-9. [DOI] [PubMed] [Google Scholar]
- 47.Ursos LM, DuBay KF, Roepe PD. Antimalarial drugs influence the pH dependent solubility of heme via apparent nucleation phenomena. Mol Biochem Parasitol. 2001;112:11–17. doi: 10.1016/s0166-6851(00)00342-x. [DOI] [PubMed] [Google Scholar]
- 48.Grabe M, Oster G. Regulation of organelle acidity. J Gen Physiol. 2001;117:329–344. doi: 10.1085/jgp.117.4.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demaurex N. pH Homeostasis of cellular organelles. News Physiol Sci. 2002;17:1–5. doi: 10.1152/physiologyonline.2002.17.1.1. [DOI] [PubMed] [Google Scholar]
- 50.Biagini GA, Fidock DA, Bray PG, Ward SA. Mutations Conferring Drug Resistance in Malaria Parasite Drug Transporters Pgh1 and PfCRT Do Not Affect Steady-State Vacuolar Ca2+ Antimicrob Agents Chemother. 2005;49:4807–4808. doi: 10.1128/AAC.49.11.4807-4808.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charbonneau M, Grey RD, Baskin R, Thomas D. A Freeze-Fracture Study of the Cortex of Xenopus laevis Eggs. Dev Growth Diff. 1986;28:75–84. doi: 10.1111/j.1440-169X.1986.00075.x. [DOI] [PubMed] [Google Scholar]