Abstract
During transfer RNA (tRNA) selection, a cognate codon:anticodon interaction triggers a series of events that ultimately results in the acceptance of that tRNA into the ribosome for peptide-bond formation. High-fidelity discrimination between the cognate tRNA and near- and noncognate ones depends both on their differential dissociation rates from the ribosome and on specific acceleration of forward rate constants by cognate species. Here we show that a mutant tRNATrp carrying a single substitution in its D-arm achieves elevated levels of miscoding by accelerating these forward rate constants independent of codon:anticodon pairing in the decoding center. These data provide evidence for a direct role for tRNA in signaling its own acceptance during decoding and support its fundamental role during the evolution of protein synthesis.
Selection of cognate aminoacyl-tRNAs (aa-tRNAs) is accomplished by the translation machinery with high accuracy and speed by means of kinetic proofreading (1, 2) and induced-fit (3, 4) mechanisms (Fig. 1). Kinetic proofreading is facilitated by the action of the guanosine triphosphatase (GTPase) elongation factor Tu (EF-Tu) in ternary complex with aa-tRNA and GTP. The GTPase activity of this complex effectively separates selection into two stages, initial selection and proofreading, which allow multiple opportunities for the rejection of incorrect tRNAs. Induced fit further increases fidelity by selectively accelerating the forward rates of two steps in the selection process—activation of EF-Tu for GTP hydrolysis (rate constant k3) and accommodation of aa-tRNA into the A site (k5)—for cognate relative to near-cognate aa-tRNAs (4, 5). This induced fit results from cognate codon:anticodon interactions somehow accelerating rate-limiting conformational changes required for GTP hydrolysis and peptidyl transfer. These changes likely originate in the decoding center of the small ribosomal subunit where the codon:anticodon interaction is “read” and are then transmitted to remote regions of the large ribosomal subunit involved in GTP hydrolysis and accommodation. Such conformational changes in the decoding center resulting from cognate (but not near-cognate) interactions have been documented in x-ray structures (6). Communication between the decoding center and the large subunit could proceed through intersubunit bridges affected by “closure” of the small subunit upon cognate tRNA binding (7–9) or through the tRNA itself (10). Here we show that the tRNA body acts as a direct functional link between the decoding center and remote regions of the ribosome that promote GTP hydrolysis and accommodation.
Fig. 1.
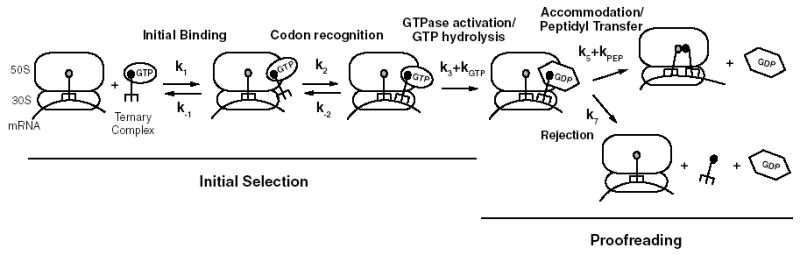
Kinetic scheme for tRNA selection on the ribosome identifying the two stages of initial selection and proofreading. The scheme includes the relevant kinetically resolved steps (4). EF-Tu is shown in different conformations in the GTP- and GDP-bound form.
The contribution of tRNA to decoding was studied by using the Hirsh suppressor (11), a tRNATrp variant that recognizes both the tryptophan (UGG) and UGA stop codons. Rather than carrying an anticodon mutation, this tRNA carries a G24A substitution in the D-arm, changing the U11:G24 base pair to U11:A24. This mutation could cause miscoding in two distinct ways: It could slow dissociation (rejection) from the ribosome [as previously argued (12)] or it could accelerate forward rate constants in tRNA selection. To distinguish between these possibilities, we first measured rate constants for two forward steps, GTPase activation (k3) and accommodation (k5), for wild-type and mutant tRNAs. Second, we addressed the effect of this mutation on dissociation of these tRNAs from the ribosome by measuring both rejection rates during proofreading (k7) and the equilibrium dissociation constant (k−2 k−1/k2k1). The pre–steady-state kinetic analysis presented here is modeled on earlier studies of Rodnina and colleagues (4, 5). Although kinetic details of tRNATrp show some variance from those previously obtained for tRNAPhe and tRNALeu, the overall view of tRNA selection that emerges is notably consistent.
We first determined rate constants for GTPase activation with wild-type and G24A tRNATrp on ribosome complexes programmed with mRNAs containing cognate (UGG) or near-cognate (UGA) codons in the A site. The GTPase activation rate (k3) was measured by following GTP hydrolysis, because activation is rate-limiting for this event (4). Rates for single-turnover GTP hydrolysis were measured by mixing programmed ribosomes with a purified ternary complex composed of EF-Tu, [γ-32P]GTP, and either wild-type or variant Trp-tRNATrp (Fig. 2A, top panel). GTP hydrolysis rates were measured at increasing ribosome concentrations, and pseudo–first-order rate constants were calculated from the extrapolation of hyperbolic fits to saturation (Fig. 2A, middle panel). Although both wild-type and G24A tRNATrp activate GTP hydrolysis to the same extent on the cognate codon (k3 = 80 s −1), the mutant tRNA increases this rate constant from ~5 to ~45 s−1 on the near-cognate codon (Fig. 2A, bottom panel, and Table 1).
Fig. 2.
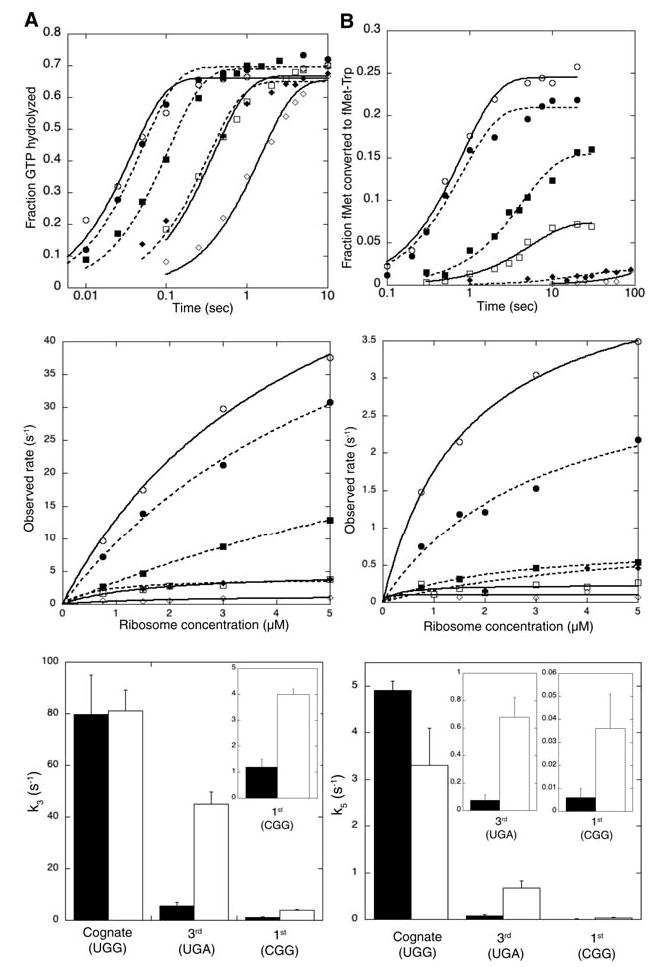
G24A tRNATrp variant accelerates forward rates of (A) GTPase activation and (B) accommodation on near-cognate codons. The top panels show representative time courses (at 2 μM ribosomes) of GTP hydrolysis and dipeptide formation for wild-type (solid lines, open symbols) and G24A (dashed lines, solid symbols) tRNATrp on cognate (circles), UGA (squares), or CGG (diamonds) programmed ribosomes. The middle panels show ribosome titrations and fits for rate-constant calculations. The bottom panels show calculated rate constants for wild-type (black) and G24A (white) tRNATrp. Each bar represents the average of two to four ribosome titration experiments and the error bars represent their standard deviations.
Table 1.
Rate and dissociation constants of wild-type and mutant tRNATrp on cognate and near-cognate codons. l.d., limit of detection; wt, wild type.
|
k3 (s−1)
|
k5 (s−1)
|
k7 (s−1)
|
Kd (nM)
|
|||||
|---|---|---|---|---|---|---|---|---|
| wt | G24A | wt | G24A | wt | G24A | wt | G24A | |
| UGG (cognate) | 79.7 ± 15.2 | 81.2 ± 8.0 | 4.9 ± 0.2 | 3.3 ± 0.8 | < l.d. | < l.d. | 1.2 ± 0.7 | 0.6 ± 0.3 |
| UGA (3rd) | 5.6 ± 1.4 | 45.1 ± 4.7 | 0.07 ± 0.04 | 0.68 ± 0.14 | 0.12 ± 0.08 | 0.19 ± 0.23 | 19.9 ± 3.5 | 25.7 ± 3.9 |
| CGG (1st) | 1.2 ± 0.3 | 4.0 ± 0.2 | 0.006 ± 0.004 | 0.036 ± 0.015 | 0.09 ± 0.06 | 0.41 ± 0.17 | 20.3 ± 4.8 | 19.3 ± 3.4 |
These data establish that the G24A substitution in the D-arm of tRNATrp bypasses the otherwise strict requirement for cognate codon:anticodon interactions to activate GTP hydrolysis. To rule out that the observed rate increase was specific for the UGA near-cognate codon, we examined whether G24A tRNATrp had similar effects on additional mRNAs [as suggested by earlier in vitro studies with UGU codons (13)]. We measured rate constants for GTPase activation (k3) on other third- (UGU and UGC) and first-position (CGG) near-cognate codons. Although the rate constants for GTPase activation are considerably lower on first-position near-cognate codons, the relative increase in k3 seen for G24A tRNATrp is similar to that observed on the third-position mismatch UGA (Fig. 2A and Table 1). Similar results were also obtained for the third-position mismatched codons UGU and UGC (14). Finally, we used a truncated mRNA to determine whether GTPase activation by mutant tRNATrp is enhanced relative to that of the wild type in the complete absence of an A-site codon. Again, G24A tRNATrp stimulates GTP hydrolysis by ~4-fold relative to that of the wild type (from 0.01 ± 0.005 to 0.04 ± 0.004 s−1, at 5 μM ribosomes) (14). The uniform stimulation of k3 by the G24A substitution, independent of codon:anticodon interaction, indicates that this miscoding tRNA variant autonomously stimulates downstream events. Moreover, it seems unlikely that the G24A substitution also overcomes deficits in the stability of the codon:anticodon interaction for such distinct mismatches, arguing against effects on k−2.
We next determined rate constants for another forward step, tRNA accommodation (k5), for wild-type and G24A tRNATrp on ribosome complexes containing cognate (UGG) or two different near-cognate (UGA and CGG) mRNAs. Here we followed peptide bond formation, because its rate (kpep) was limited by accommodation (k5) (4). After the mixing of programmed ribosomes containing f-[35S]-Met-tRNAfMet in the P site with wild-type and mutant Trp-tRNATrp ternary complex, the amount of fMet-Trp dipeptide formed over time was quantitated (Fig. 2B, top panel). Because accommodation (k5) is at a branch point in the kinetic pathway (Fig. 1), the observed rate constants (kobs) obtained from ribosome titrations (Fig. 2B, middle panel) represent the sum of the individual rate constants of accommodation and rejection (kobs = k5 + k7). For cognate interactions, rejection is negligible (4, 5), and thus the observed rate constant simply represents accommodation (kobs = k5). For near-cognate interactions, the extent of dipeptide formation is substantially reduced (Fig. 2B, top panel), indicating that a fraction of bound tRNA proceeds through the productive pathway, whereas the rest is irreversibly rejected [fraction = k5/(k5 + k7)]. Because accommodation is rate-limiting, we can calculate individual rate constants (k5 and k7) for near-cognate complexes. As observed for k3, rate constants for accommodation (k5) are similar for wild-type and mutant tRNATrp on the cognate codon. On near-cognate codons, however, G24A tRNATrp increases accommodation rates by six- to ninefold relative to wild type (Fig. 2B, bottom panel and Table 1). Thus, variant tRNATrp promotes general miscoding by accelerating the other forward step, k5, in tRNA selection.
The observed stimulation of forward rate constants (k3 and k5) by G24A tRNATrp establishes that miscoding is promoted by kinetic contributions to induced fit. To investigate whether this mutation also increases miscoding by slowing dissociation, we examined rate constants of rejection during proofreading (k7) (Fig. 3A and Table 1). The rejection rate for G24A tRNATrp is equal to or higher than that for the wild type, indicating that increased residence time on the ribosome of the variant tRNA does not contribute to its miscoding capacity.
Fig. 3.
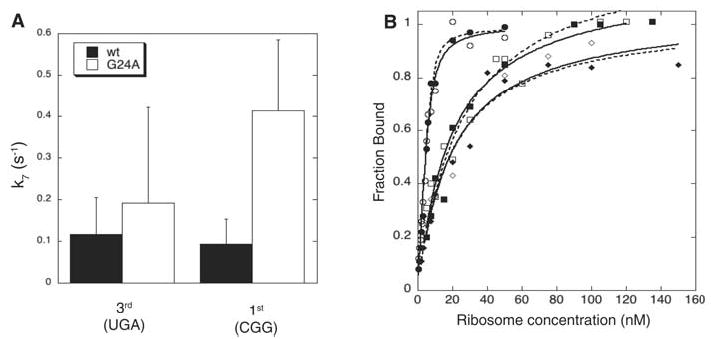
G24A tRNATrp variant does not increase miscoding levels by slowing rejection from the ribosome. (A) Rate constants of rejection during proofreading (k7) were calculated from the rate and extent of dipeptide formation for wild-type (wt) (black) and G24A (white) tRNATrp. Errors were calculated by standard error propagation. (B) Equilibrium dissociation constants (Kd’s) were measured by filter-binding for wild-type (solid lines, open symbols) and G24A tRNA (dashed lines, solid symbols) on cognate (circles), UGA (squares), or CGG (diamonds) programmed ribosomes.
To examine the other dissociation step, k−2, we measured equilibrium dissociation constants (Kd = k−1k−2/k1k2) between tRNAs in ternary complexes with a GTPase-deficient EF-Tu variant (15) and cognate- or near-cognate programmed ribosomes. The Kd’s measured by filter binding were similar for wild-type and G24A tRNATrp ternary complexes on each programmed ribosome (Fig. 3B and Table 1). This observation implies that the dissociation rate constant, k−2, is un-affected by the G24A substitution in tRNATrp, because otherwise changes in k−2 would need to be exactly compensated by changes in the other rate constants (k1, k2, and k−1). Because k1, k−1, and k2 are indistinguishable for different tRNA species on various cognate and near-cognate codons (4, 5), this compensation seems unlikely.
Our data establish that the D-arm substitution stimulates miscoding through acceleration of forward selection rates. Thus, the miscoding tRNA variant takes advantage of the induced-fit mechanism for tRNA selection, and in doing so, triggers downstream events, bypassing normally required signals from the decoding center. The two rate constants, k3 and k5, are similarly affected, suggesting that a common property of tRNA is central to the mechanism of both steps. These results do not preclude a role for the ribosome itself in the signaling process.
The D-arm substitution likely has a direct effect on tRNA deformability, somehow facilitating required conformational changes during decoding. Altered flexibility of G24A tRNATrp is supported by earlier work, showing that variant tRNATrp exhibits slower rates of intramolecular cross-linking (16). The cross-linked G24A tRNATrp no longer suppresses UGA stop codons but is still otherwise functional (17). These observations suggest that flexibility in tRNA plays a critical role in decoding.
Cryo–electron microscopy studies provide a view of tRNA in a pre-accommodated state (18) that reveals a “kink” between the anticodon- and D-stems, close to position G24 of tRNA, providing evidence that tRNA structural conformers are relevant intermediates in decoding. Alterations of tRNA in this region might affect its capacity to assume this kinked conformation and thus promote downstream events (19). It also is possible that G24A tRNATrp interacts differently with nearby ribosomal elements including helix 69, the sarcin-ricin loop, and the GTPase associated center, all candidates for participating in signal transmission during tRNA selection (18, 20, 21).
Our studies suggest that conformational changes in tRNA are the physical basis for induced fit, which is an essential contributor to high-fidelity tRNA selection (5). Protein synthesis is widely thought to have evolved in an RNA-dominated world where, in the absence of sophisticated factors like EF-Tu, the earliest translational machinery must have relied on active contributions from tRNA. Our results mesh well with recent studies detailing an active role for tRNA in peptidyl transfer (22). The recognition of tRNA as an active player in translation (23) will replace the historical view of tRNA as a static “adaptor” as more specific roles for tRNA are uncovered.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/308/5725/1178/
Materials and Methods
References
References
- 1.Ninio J. Biochimie. 1975;57:587. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 2.Hopfield JJ. Proc Natl Acad Sci USA. 1974;71:4135. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koshland DE. Proc Natl Acad Sci USA. 1958;44:98. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pape T, Wintermeyer W, Rodnina MV. EMBO J. 1999;18:3800. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gromadski KB, Rodnina MV. Mol Cell. 2004;13:191. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 6.Ogle JM, et al. Science. 2001;292:897. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 7.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Cell. 2002;111:721. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 8.Gabashvili IS, et al. Cell. 2000;100:537. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 9.Yusupov MM, et al. Science. 2001;292:883. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 10.Piepenburg O, et al. Biochemistry. 2000;39:1734. doi: 10.1021/bi992331y. [DOI] [PubMed] [Google Scholar]
- 11.Hirsh D. J Mol Biol. 1971;58:439. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- 12.Smith D, Yarus M. J Mol Biol. 1989;206:489. doi: 10.1016/0022-2836(89)90496-8. [DOI] [PubMed] [Google Scholar]
- 13.Buckingham RH, Kurland CG. Proc Natl Acad Sci USA. 1977;74:5496. doi: 10.1073/pnas.74.12.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.L. Cochella, R. Green, unpublished data.
- 15.Daviter T, Wieden HJ, Rodnina MV. J Mol Biol. 2003;332:689. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- 16.Favre A, Buchingham R, Thomas G. Nucleic Acids Res. 1975;2:1421. doi: 10.1093/nar/2.8.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vacher J, Buckingham RH. J Mol Biol. 1979;129:287. doi: 10.1016/0022-2836(79)90282-1. [DOI] [PubMed] [Google Scholar]
- 18.Valle M, et al. Nat Struct Biol. 2003;10:899. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 19.Ogle JM, Ramakrishnan V. Annu Rev Biochem2004 doi: 10.1146/annurev.biochem.74.061903.155440. 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 20.Moazed D, Robertson JM, Noller HF. Nature. 1988;334:362. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 21.Stark H, et al. Nat Struct Biol. 2002;9:849. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 22.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Nat Struct Mol Biol. 2004;11:1101. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 23.Woese CR. RNA. 2001;7:1055. doi: 10.1017/s1355838201010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.We thank M. Rodnina for advice; O. Uhlenbeck and M. Yarus for reagents; and J. Lorsch, C. Merryman, and R. Reed for comments. R.G. is an investigator of the Howard Hughes Medical Institute. This work was supported by NIH grant R01GM059425.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


