Abstract
The ubiquitin-binding RPN-10 protein serves as a ubiquitin receptor that delivers client proteins to the 26S proteasome. Although ubiquitin recognition is an essential step for proteasomal destruction, deletion of the rpn-10 gene in yeast does not influence viability, indicating redundancy of the substrate delivery pathway. However, their specificity and biological relevance in higher eukaryotes is still enigmatic. We report herein that knockdown of the rpn-10 gene, but not any other proteasome subunit genes, sexually transforms hermaphrodites to females by eliminating hermaphrodite spermatogenesis in Caenorhabditis elegans. The feminization phenotype induced by deletion of the rpn-10 gene was rescued by knockdown of tra-2, one of sexual fate decision genes promoting female development, and its downstream target tra-1, indicating that the TRA-2–mediated sex determination pathway is crucial for the Δrpn-10–induced sterile phenotype. Intriguingly, we found that co-knockdown of rpn-10 and functionally related ubiquitin ligase ufd-2 overcomes the germline-musculinizing effect of fem-3(gf). Furthermore, TRA-2 proteins accumulated in rpn-10-defective worms. Our results show that the RPN-10–mediated ubiquitin pathway is indispensable for control of the TRA-2–mediated sex-determining pathway.
INTRODUCTION
Ubiquitin is a covalent modifier that produces a polyubiquitin chain functioning as a degradation signal (Hershko and Ciechanover, 1998; Pickart, 2001). Recruitment of polyubiquitinated proteins to the degradation machinery is a key step in the selective degradation of various cellular proteins (Pickart, 1998; Voges et al., 1999; Madura, 2004). Polyubiquitin chains with lengths of at least four ubiquitin moieties can be recognized and degraded by the 26S proteasome, a eukaryotic ATP-dependent protease complex (Voges et al., 1999; Thrower et al., 2000). The 26S proteasome is composed of the catalytic 20S proteasome and the regulatory PA700 complex; the later complex consists of six ATPase subunits (RPT-1-6) and multiple non-ATPase subunits (RPN-1 -3, -5-15), each ranging in size from 11 to 110 kDa (Tanaka, 1998).
Previous studies have shown that the RPN-10 subunit of PA700, originally called S5a, can bind to a polyubiquitin chain linked to proteins in vitro and in vivo, and it is thought to play a role as a ubiquitin receptor of the 26S proteasome (Deveraux et al., 1994; Ferrell et al., 1996; van Nocker et al., 1996; Kawahara et al., 2000; Wilkinson et al., 2001; Elsasser et al., 2004; Verma et al., 2004). Figure 1 shows an abbreviated model that highlights the RPN-10 ubiquitin-recognition subunits in 26S proteasomes. The RPN-10 has at least two distinct domains: one domain is the 60-residue N-terminal called the VWA domain, which was reported to be involved in the integration of the 26S complex (Glickman et al., 1998; Verma et al., 2004), and the other domain is the C-terminal region containing two independent polyubiquitin-binding sites, named ubiquitin interacting motif (UIM)1 and UIM2 (Young et al., 1998; Hofmann and Falquet, 2001). Although it was thought that ubiquitin recognition is an essential step in the proteasome-mediated degradation process, the role of the RPN-10 subunit as a ubiquitin receptor of the 26S proteasome has become a subject of debate after the observation that its deletion in yeast does not influence the viability of the cells and causes only a mild phenotype with increased steady-state levels of ubiquitin–protein conjugates (van Nocker et al., 1996; Fu et al., 1998; Szlanka et al., 2002). This observation suggested the existence of other different ubiquitin recognition mechanism(s). Indeed, several proteins such as Rad23p and Dsk2p are involved in polyubiquitinated substrate delivery to the proteasome in yeast, because they have a ubiquitin-associated domain (UBA) that can bind to polyubiquitin chains and a ubiquitin-like domain capable of associating with the 26S proteasome subunits (Wilkinson et al., 2001; Saeki et al., 2002; Elsasser et al., 2004). Accordingly, these gene products in the substrate delivery pathway may play a largely redundant role. However, their specific target proteins and biological relevance in higher eukaryotes remain a mystery to be uncovered.
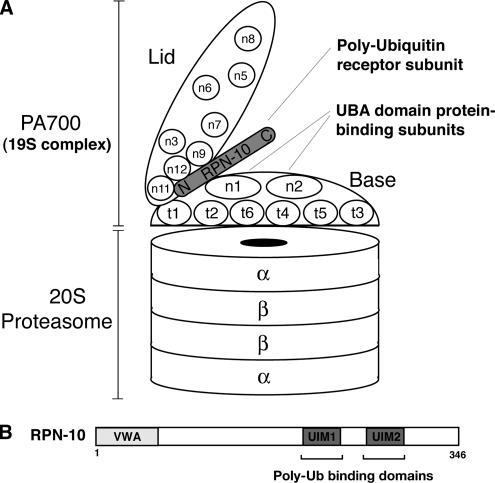
Figure 1.
(A) Schematic representation of the 26S proteasome. The 26S proteasome is composed of the catalytic 20S proteasome and a regulatory particle termed PA700, which can be subdivided into the Base and the Lid complex. RPN-10 is an intrinsic ubiquitin receptor of 26S proteasomes and is highlighted in the figure. It was also reported that there are alternative ubiquitin receptors of 26S proteasome, UBL-UBA domain proteins, and that they interact with RPN-1 and possibly RPN-2 subunits of the Base complex of 26S proteasome. (B) Schematic diagram of RPN-10 ubiquitin receptor. Previous studies have shown that RPN-10 can bind to the Base complex via N-terminal VWA domain and bind to polyubiquitinated substrates via its C-terminal domain, including polyubiquitin (poly-Ub)-binding UIMs.
The genetic basis of sex determination has been analyzed extensively in the nematode, which has two naturally occurring sexes, an XX hermaphrodite and an XO male (Kuwabara and Kimble, 1992; Meyer, 2000; Kuwabara and Perry, 2001; Puoti et al., 2001). Hermaphrodites sequentially produce sperm and oocytes from a single pool of precursors. After progression of L4 to adult molt, spermatogenesis is turned off, and all of the gametes that form subsequently develop into oocytes. Most of the sexual fate decision genes have been identified and have been shown to function in a cascade of negative regulation (Ellis and Schedl, 2006; Zarkower, 2006). Sex determination requires posttranscriptional regulation of two key genes, tra-2 and fem-3. The balance between the activities of TRA-2 and FEM-3 influences the decision to adopt either a female or male fate. Genetic studies have shown that tra-2 promotes female development because loss-of-function mutations of tra-2 transform XX animals into pseudomales that do not produce oocytes (Hodgkin and Brenner, 1977), whereas tra-2 eg (extra gain-of-function) mutant allele lead to dominant transformation of XO animal from fertile male into fertile female (Hodgkin and Albertson, 1995; Kuwabara, 1996). TRA-2 is a large transmembrane protein that somewhat resembles the Patched receptor (Kuwabara and Kimble, 1992; Kuwabara, 1996), and it was thought that the potency of TRA-2 is regulated by a variety of mechanisms; it is controlled by its ligand HER-1 (Perry et al., 1993; Kuwabara, 1996), by TRA-1–mediated nuclear export of tra-2 mRNA (Graves et al., 1999; Segal et al., 2001) and by translational regulation through 3′-untranslated region (UTR)-binding proteins (Goodwin et al., 1997; Clifford et al., 2000; Puoti et al., 2001). It has recently been suggested that the C-terminal intracellular domain (ICD) of TRA-2 is likely to be produced as a soluble fragment in vivo through proteolytic cleavage of TRA-2 (Sokol and Kuwabara, 2000) or through translation of an XX germline-specific alternative transcript (Kuwabara et al., 1998). It has been shown that the heat-shock promoter-induced exogenous intracellular domain of TRA-2 interacts with global sex regulatory transcriptional factor TRA-1 in the nuclei of both XX and XO animals (Lum et al., 2000; Wang and Kimble, 2001) and that this fragment has feminizing activity (Kuwabara and Kimble 1995). The intracellular domain of TRA-2 also interacts with FEM proteins and modulates their activity (Mehra et al., 1999). Both FEM-1 and FEM-3 are targeted for proteasomal degradation by the F-box ubiquitin ligase SEL-10 (Jager et al., 2004). However, there have been only a few studies on the regulation of sex determination by the ubiquitin-mediated proteolytic pathway (Clifford et al., 2000; Jager et al., 2004). To investigate the function of the ubiquitin receptors of the 26S proteasome in higher eukaryotes, we focused on Caenorhabditis elegans counterpart of RPN-10. Our results clearly show the biological requirement of the RPN-10–mediated substrate discrimination system that is indispensable for the control of the sex-determining signaling pathway.
MATERIALS AND METHODS
Nematode Strains
C. elegans strains were cultured using standard techniques (Brenner, 1974). C. elegans N2 variety Bristol was used for RNA interference (RNAi) analysis and for wild-type observations. The rpn-10 mutant alleles tm1180 and tm1349 was isolated by the National Bioresource Project for the Nematode (NBPN; Department of Physiology, School of Medicine, Tokyo Women's Medical University, Tokyo, Japan). Both rpn-10 (tm1180) and rpn-10 (tm1349) alleles, generous gifts from Dr. S. Mitani (NBPN), were backcrossed at least five times against wild-type males before analysis. Although the original tm1349 strain from National Bioresource Project shows only modest defects in the production of progeny, we found that repeated rounds of backcross against wild-type males resulted in generation of sterile females, and we used a backcrossed strain as rpn-10 (tm1349) in this study unless otherwise noted. We have confirmed that further backcross procedures did not influence the penetrance of rpn-10 (tm1349) phenotype and that this mutant did not express the corresponding transcript. fog-2(oz40), fem-2(b245ts), tra-2 (e1095), tra-2 (e2020), fem-3 (q20), and fem-3 (e1996) mutant worms were kindly provided by Dr. T. Schedl (Washington University, St. Louis, MO) and the Caenorhabditis elegans Genetics Center (University of Minnesota, St. Paul, MN), respectively.
RNA Interference
RNAi experiments were mainly performed by the method of feeding (Timmons et al., 2001), and the results were confirmed by the injection method (Fire et al., 1998; Shimada et al., 2002). Efficacy of gene expression knockdown in each of RNAi experiments were verified by reverse transcription-polymerase chain reaction (RT-PCR) analysis. rpn-10 (B0205.3), ufd-2 (T05H10.5), and other cloned genes (rpn-1, T22D1.9; tra-1, Y47D3A.6; tra-2, C15F1.3; oma-1, C09G9.6; oma-2, ZC513.6; rad-23, ZK20.3; and dsk-2, F15C11.2) were amplified by PCR from a C. elegans mixed stage cDNA library. Primer sequences are available from the author on request. The amplified PCR products were subcloned into a pPD129.36 vector and transfected into Escherichia coli strain HT115 (DE3). RNA transcription from pPD129.36 vector plasmids was induced by the addition of 0.4 mM isopropyl β-d-thiogalactoside (IPTG) to E. coli culture medium (at an optical density of 0.4), followed by incubation for 4 h at 37°C. The harvested E. coli was spread on NGM plates (containing 50 μg/ml ampicillin, 12.5 μg/ml tetracycline, and 0.4 mM IPTG) for the feeding experiments. Feeding was started at the L1 stage, and worms were cultured for the indicated times at 25°C unless otherwise noted. To show self-sterility, worms were picked at the L4/young adult stage to individual plates and assayed for their ability to produce progeny.
Glycerol Density Gradient Centrifugation
For sedimentation velocity analysis, worms were homogenized with 26S proteasome extraction buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 2 mM MgCl2, 1 mM dithiothreitol, 2 mM ATP, and 10% glycerol) and centrifuged at 10,000 × g for 20 min to obtain soluble extracts. Extracts of 5 mg of protein were subjected to glycerol density-gradient centrifugation with 10–40% glycerol. After centrifugation at 83,000 × g for 22 h, the gradient was separated into 33 fractions of 1 ml each. The activity of the 26S proteasome in the fractions was measured by using the synthetic peptide succinyl-Leu-Leu-Val-Tyr-4-methylcoumarin-7-amide (Suc-LLVY-MCA) as described previously (Kawahara et al., 2000). Purified mouse 20S and 26S proteasomes were used as markers for sedimentation.
Examination of Spermatheca
To examine the state of the spermatheca, adult hermaphrodites (or females) were cut open to release gonads, and freeze/cracked samples in M9 buffer were fixed by incubation in cold methanol (−20°C) followed by cold acetone (−20°C). Fixed gonads were stained with Hoechst 33342 at the concentration of 2.5 μg/ml.
Mating with Males
Individual F1 sterile worms of rpn-10;ufd-2 (RNAi) and rpn-10 (tm1349) homozygote at L4 stage were each plated with each of six healthy young-adult males and crossed at 20°C. At 24 h after crossing, parental worms were removed from the plates. After another 24-h incubation, the number of progeny was counted. Oocyte-defective oma-1;oma-2 (RNAi) worms and feminized fem-2 (b245ts) mutant worms were crossed with males under conditions similar to those described above and used as negative and positive controls, respectively, for this experiment.
Effect of RNAi on XO Males
XO males (P0) were crossed with XX hermaphrodites (P0) treated with either rpn-10;ufd-2 (RNAi)- or control vector (RNAi), and resulting F1 larvae were continued to be treated with respective RNAi until the young adult stage at 20°C. After verification of F1 feminization effects in rpn-10;ufd-2 (RNAi)-treated hermaphrodites, individual F1 males were harvested and subjected to either microscopic observations or RT-PCR analyses.
Immunological Analysis
The anti-TRA-2 ICD antibody was prepared for this study as follows. Five hundred micrograms of bacterially produced TRA-2 intracellular domain (amino acids 1135–1475 of C. elegans TRA-2 protein) was mixed and emulsified with an equal amount of TiterMaxGold (TiterMax USA, Norcross, GA) and then inoculated into a rabbit. The antibody was obtained after six rounds of immunization at 2-wk intervals and used after affinity purification.
Indirect immunofluorescence analyses were performed as reported by Finney and Ruvkun (1990) with modifications as follows. Animals were suspended in 4% paraformaldehyde, 25% methanol, and quick-frozen in liquid nitrogen. After thawing on ice, the animals were incubated on ice for 1 h to complete fixing, washed three times in 1% Triton X-100, 100 mM Tris-HCl, pH 7.5, and incubated in 1% Triton X-100, 100 mM Tris-HCl, pH 7.5, and 1% β-mercaptoethanol for 2 h at 37°C to reduce the cuticle. After three washes in 10 mM Na BO3, pH 9.2, the animals were incubated in 10 mM Na BO3, pH 9.2, and 0.3% H2O2 for 1 h at 25°C. The animals were then washed two times in 10 mM Na BO3, pH 9.2, and stored in PTB buffer (1X phosphate-buffered saline [PBS], 0.1% Triton X-100, 0.1% bovine serum albumin, and 0.05% NaN3). After blocking with blocking solution (1X PBS, 0.1% Tween 20, and 5% bovine serum albumin) for 30 min at 25°C, antibody incubations were performed overnight at 4°C in the blocking solution. Affinity-purified anti-TRA-2 ICD antibody was diluted to a concentration of 1 μg/ml before incubation. After three washes in 1× PBS and 0.1% Tween 20 (PBS-T), the worms were incubated for 90 min at 25°C with 1:800 Alexa 488-conjugated anti-rabbit IgG (Invitrogen, Carlsbad, CA) in blocking solution. After four washes in PBS-T, the worms were treated with 2.5 μg/ml Hoechst 33342 for 15 min at 25°C and then washed another three times in PBS-T. Then, small aliquots of worms were mounted on slides with VECTASHIELD (Vector Laboratories, Burlingame, CA) mounting media and examined under an Axioplan II microscope (Carl Zeiss, Jena, Germany) equipped with fluorescence capabilities.
For general immunoblot analyses, extracts from RNAi-treated and mutant worms or cultured cell extracts were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). The membranes were probed with an anti-RPN-10 peptide antibody (37CHSKTRSNPENNVGLITLAN56 as an antigen for C. elegans RPN-10), anti-ubiquitin antibody, anti-20S proteasome C2 subunit antibody, anti-tubulin DM1a monoclonal antibody (Sigma-Aldrich, St. Louis, MO), and anti-FLAG M2 antibody (Sigma-Aldrich).
For anti-TRA-2 ICD blots, samples were prepared by picking up 50 appropriate animals, boiling in 2X SDS-PAGE sample buffer with 1% β-mercaptoethanol, separating by SDS-PAGE, and transferring onto Immobilon P transfer membranes (Millipore, Billerica, MA). Primary antibody for anti-TRA-2 ICD was used at a concentration of 0.2 μg/ml in reaction buffer (1X PBS, 5% skim milk, 3% normal goat serum, 3% E. coli HT115 extract, and 0.1% Tween 20). Biotinylated goat anti-rabbit IgG and Vectastain ABC detection reagents (Vector Laboratories) were used, followed by detection with ECL Western blotting detection reagents (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
RESULTS
Knockdown of RPN-10 Results in Induction of F1 Sterility
Although reduction of the expression of most proteasomal subunits by RNAi results in immediate growth arrest in C. elegans, we found that inhibition of rpn-10 gene expression did not cause any obvious defects in P0 growth, reproduction, and subsequent development of F1 embryos (Figure 2, A and B). Because mutation in the ufd-2 gene, which was originally identified by genetic dissection of the UFD pathway (Johnson et al., 1995), with rpn-10 shows synthetic growth defects in yeast (Koegl et al., 1999), we attempted to inhibit rpn-10 and ufd-2 gene expressions simultaneously. Again, we were unable to detect any obvious defects during P0 generation (Figure 2B). Efficacy of gene expression knockdown in each of the RNAi experiments was verified by RT-PCR analysis, confirming that these genes seem to be dispensable for these stages. In rpn-10;ufd-2 (RNAi) worms, peptidase activity, i.e., chymotryptic Suc-LLVY-MCA-degrading activity, of the 26S proteasome and its sedimentation profile was indistinguishable from that in wild-type worms (Figure 2D), despite the reduction of rpn-10 gene expression (Figure 2, E and F). These observations indicate that the overall 26S proteasome architecture and functions for cell viability are intact in the rpn-10- and ufd-2–defective environment. However, we noticed the accumulation of polyubiquitinated proteins in the extract of rpn-10 (RNAi) as well as rpn-10;ufd-2 (RNAi) worms (Figure 2G), suggesting certain defects of degradation of polyubiquitinated proteins in these RPN-10-lacking worms without affecting viability.
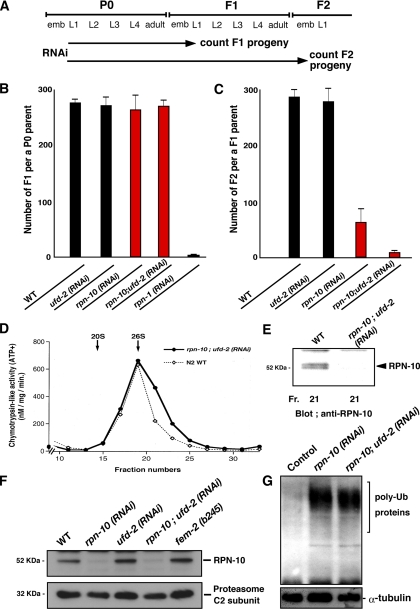
Figure 2.
RPN-10 knockdown results in F1 sterility in C. elegans. (A) Schematic diagram of RNAi treatment. (B) Effects of RNAi on the reproduction of P0 parents. The average numbers of F1 progeny per a P0 parent are indicated. rpn-1 (RNAi) was used as a positive control for this RNAi experiment. (C) Effects of RNAi on the reproduction of F1 parents. The average numbers of F2 progeny per an F1 parent are indicated. Note that double RNAi treatment of rpn-10 and ufd-2 results in a nearly complete sterile phenotype. (D) Sedimentation velocity analysis of the 26S proteasome. Worm extracts were fractionated by glycerol density-gradient centrifugation and assayed for 26S proteasomal chymotrypsin-like activity. Elution points of 20S and 26S proteasomes are indicated by arrows. (E) Proteins in the 26S proteasome fraction (Fr. 21) were precipitated with acetone and subjected to immunoblotting with anti-RPN-10 antibody. (F) Expression of RPN-10 protein is suppressed in rpn-10 (RNAi) worms. Immunosignal of the 20S proteasome C2 subunit was used as a control. (G) RPN-10 knockdown results in accumulation of polyubiquitinated (poly-Ub) proteins. α-tubulin as a loading control.
Although there was no apparent defect at P0 stage in rpn-10-knockdown worms, close inspection revealed a prominent reduction in the number of F2 embryos laid from rpn-10 (RNAi) F1 worms (Figure 2C). Inhibition of ufd-2 expression alone resulted in an essentially no phenotype in F1 and later stage worms, but double inhibition of rpn-10 and ufd-2 expression resulted in a further additional reduction of the number of F2 eggs laid, i.e., >97% reduction in the number of F2 progeny compared with the control (Figure 2C). These observations suggested that the rpn-10 gene possess important functions for the production of F2 progeny in collaboration with ufd-2 gene and that their removal results in a sterile phenotype in F1 stage worms.
Knockdown of RPN-10 Results in a Defect in Sperm Formation
To determine the defect that leads to F1 sterility, we first examined the F1 phenotypes in detail. In wild-type worms, germ cells proliferated by repeated rounds of mitosis on the distal side of the gonad and then progressed into meiosis and moved proximally, producing maturing oocytes on the proximal side. We found that the proximal gonad arm expands greatly in rpn-10–inhibited F1 individuals (Figure 3C) compared with that in wild-type gonads (Figure 3A), and germinal vesicle breakdown in proximal oocytes is likely to occur at a very low level. The double RNAi of rpn-10 and ufd-2 shows more penetrance but essentially the same phenotype as that of rpn-10–compromised individuals (Figure 3B), whereas single RNAi against ufd-2 induced no abnormality in the gonad (Figure 3D).
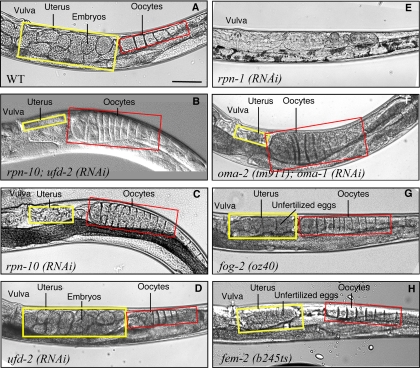
Figure 3.
RPN-10 knockdown induces characteristic reproductive defects. Compared with the WT gonad (A), typical rpn-10 (RNAi) worms show expanded oocytes (indicated by a red box) and nearly empty uterus (indicated by a yellow box) (C), representing similar morphology to oocytes-defective oma-2 (tm911);oma-1 (RNAi) worms (F). Double RNAi of rpn-10 and ufd-2 results in more penetration but essentially the same phenotype as in C (B). ufd-2 (RNAi) alone shows no abnormality in gonads (D). Knockdown of the expression of RPN-1, a constitutive subunit of the 26S proteasome, induces immediate growth arrest (E). Feminized fog-2(oz40) (G) and fem-2(b245ts) mutant (H), show accumulation of unfertilized oocytes in the uterus due to their stochastic maturation. Images on the left-top side correspond to the ventral uterus. Bar, 50 μm.
To check whether the sterile but viable phenotype of rpn-10 (RNAi) is caused by partial retardation of general 26S proteasome ability, we examined the effects of continuous reduction of 26S proteasome function. Efficient knockdown of the expression of RPN-1, a constitutive and essential subunit of the 26S proteasome, resulted in immediate arrest of larval growth and/or extensive death (Figure 3E). Continuous knockdown of rpn-1 by modest feeding of double-stranded RNA to C. elegans hermaphrodites can result in partial depletion of the product of this gene (Kamath et al., 2001 and Supplemental Table SI). Irrespective of any extent of partial inhibition of rpn-1, no individuals showed similar reproductive defects as seen in rpn-10 (RNAi) (Supplemental Figure S1). Furthermore, there was no synthetic effect between ufd-2 (RNAi) with partial knockdown of rpn-1 (Supplemental Table SII). Thus, it was concluded that the phenotype of rpn-10 (RNAi) is not due to general retardation of proteasome activity.
Oocyte maturation defects in C. elegans so far reported are feminizing mutations such as fem-2 (b245ts) and fog-2 (oz40) (Figure 3, G and H; Schedl and Kimble, 1988; McCarter et al., 1999; Miller et al., 2001, 2003) as well as defects in oocyte-specific oma-family genes (Figure 3F; Detwiler et al., 2001; Shimada et al., 2002, 2006). To determine whether the defects in rpn-10–compromised individuals are due to an abnormality in oocyte or sperm, we performed mating experiments with normal males and sterile worms. After being crossed with wild-type (N2) males, fem-2 (b245ts) mutant females were able to produce progeny, whereas oocyte-defective oma-1;oma-2 (RNAi) worms were not rescued by the wild-type male sperm (Figure 4A). It was found that rpn-10;ufd-2 (RNAi) sterile worms can be cross-fertilized by wild-type males to produce normal viable F2 embryos (Figure 4A). These findings support the view that the primary defect induced by rpn-10;ufd-2 (RNAi) is likely to be caused by a defect in the sperm.
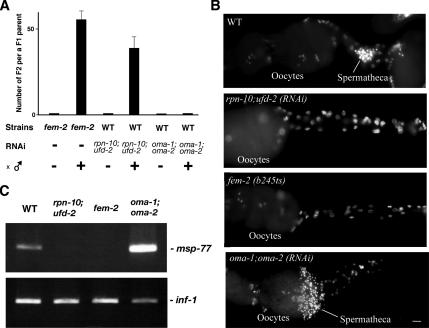
Figure 4.
The primary defect in RPN-10-knockdown worm is sperm formation. (A) Mating experiments with normal males and sterile worms. With excess males, feminized fem-2(b245ts) mutants were able to produce progeny, whereas oocyte-defective oma-1;oma-2 (RNAi) worms were not. rpn-10;ufd-2 (RNAi) sterile worms can be cross-fertilized by wild-type (N2) males to produce normal viable F2 embryos. (B) Hoechst staining of dissected gonads from WT, rpn-10;ufd-2 (RNAi), fem-2(b245ts), and oma-1;oma-2 (RNAi) worms. A number of sperm signals are evident in WT and oma-1;oma-2 (RNAi) worms as indicated but are absent in fem-2(b245ts) and rpn-10;ufd-2 (RNAi) worms. Bar, 10 μm. (C) RT-PCR analysis of the sperm-specific marker msp-77. Transcript of msp-77 is not expressed in fem-2(b245ts) and rpn-10;ufd-2 (RNAi) worms. inf-1 transcript (encoding translation initiation factor CeIF) amplified under similar conditions was used as a control.
To demonstrate directly the presence of defective sperm production, we dissected gonads from worms and stained them with Hoechst. In wild-type (WT) and oma-1;oma-2 (RNAi) hermaphrodites, many sperm were present in the spermatheca, whereas the sperm was completely absent in the spermatheca of rpn-10;ufd-2 (RNAi) worms, as in feminized fem-2(b245ts) worms (Figure 4B). As noted later using rpn-10 null mutant strain, primary and secondary spermatocytes were absent in these feminized gonads at late L4 or young adult stages (data not shown). Consistent with these morphological observations, RT-PCR quantification of the sperm-specific marker msp-77 clearly showed that sperm were in fact absent in rpn-10;ufd-2 (RNAi) worms as in the fem-2(b245ts) female (Figure 4C). These results indicate that the primary defect in rpn-10;ufd-2 (RNAi) worms is in the production of sperm, and sterility induced by reduction of the RPN-10–mediated protein degradation pathway could be explained as a consequence of feminization.
rpn-10 Mutant Worms Show Feminization Phenotype
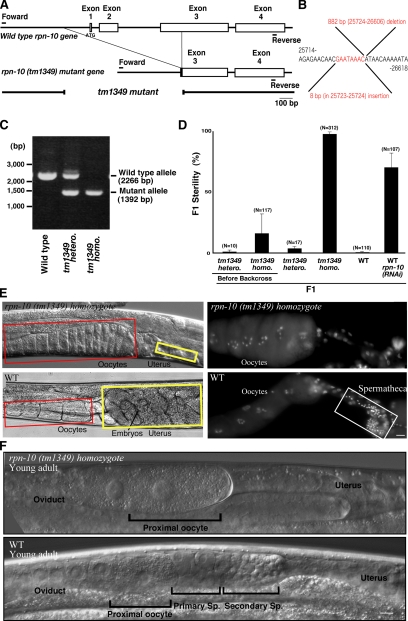
tm1349 is a null rpn-10 allele that includes a 882-base pair deletion and an 8-base pair insertion of the rpn-10 gene locus (Figure 5, A–C), resulting in loss of the first three exons of the rpn-10 gene (Figure 5A). As shown in Figure 5D, adequately backcrossed rpn-10 (tm1349) homozygotes show nearly complete F1 sterility. The morphological phenotype of rpn-10 (tm1349) homozygote is identical to that of rpn-10;ufd-2 (RNAi) worms: essentially no sperm in the spermatheca, no embryos in the uterus, and characteristic expansion of the proximal oocytes at a relatively young adult stage (54 h from L1 at 25°C) (Figure 5E; our unpublished observations). Furthermore, we found that primary and secondary spermatocytes were absent in rpn-10 (tm1349) homozygote gonads at young adult stages having only one or at most two oocytes (Figure 5F). As in rpn-10;ufd-2 (RNAi) worms, exogenously supplied sperm restored fertility; an average of 155 progeny were produced from an rpn-10 (tm1349) homozygote female that was crossed with wild-type males. These data further support the idea that the rpn-10 (tm1349) homozygote has defects in sperm production.
Figure 5.
tm1349, a null allele of rpn-10, shows a feminized phenotype. (A) Schematic diagram of wild-type and tm1349 mutant allele of the rpn-10 gene. C. elegans rpn-10 gene is composed of four exons, and the tm1349 mutant completely lacked the first and second exons with partial deletion of the third exon. Boxes, exons; lines, introns. Extent of the 882-base pair rpn-10 (tm1349) deletion is indicated by a gap. (B) Nucleotide sequence of the mutated region of the rpn-10 gene in tm1349 allele. rpn-10 cDNA (genome) obtained from a tm1349 homozygote was sequenced, and a 882-base pair deletion and 8-base pair insertion were identified in the region indicated. No sequence difference was identified around this region before and after backcrossing. The numbers of the sequence denote the wildtype genome nucleotide number of chromosome I. (C) Genomic PCR analysis confirming deletion of the rpn-10 gene in rpn-10 (tm1349) heterozygote and homozygote worms. (D) The average F1 sterilities at 25°C are indicated. Note that F1 of the rpn-10 (tm1349) homozygote shows nearly complete sterility. (E) rpn-10 (tm1349) mutant adult worms show expanded oocytes (indicated by a red box) and a nearly empty uterus (indicated by a yellow box). Hoechst staining of dissected gonads shows essentially no sperm in rpn-10 (tm1349). Spermatheca is indicated by a white box. Photographs were taken at the adult stage (54 h from L1 at 25°C). (F) Nomarski observations of gametogenesis at the young adult stage of an rpn-10 (tm1349) homozygote worm. Proximal oocytes and primary and secondary spermatocytes (labeled “Primary Sp.” and “Secondary Sp.,” respectively) are indicated. Bar, 10 μm.
RPN-10 protein is comprised of the N-terminal VWA domain and the C-terminal domain containing UIMs (Figure 1). In yeast, the modest stress-sensitive phenotype of an RPN10 deletion mutant is completely rescued by the N-terminal fragment of Rpn10p, indicating that the C-terminal domain is dispensable (Fu et al., 1998). To determine whether the C-terminal domain of C. elegans RPN-10 is essential for the feminization phenotype, we used another mutant strain rpn-10 (tm1180) (kindly provided by Dr. S. Mitani). In this mutant, exon 3 of rpn-10 gene was disrupted with a 356-base pair deletion and the gene product was expressed as a C-terminal truncated form (Supplemental Figure S2B, left, and F). As a result, polyubiquitinated proteins accumulated in rpn-10 (tm1180) worms (Supplemental Figure S2B, right) as seen in rpn-10 (RNAi) worms (Figure 2E). The rpn-10 (tm1180) is a hypomorphic allele that shows modest impairment of progeny production (Supplemental Figure S2C), but it also represents a synthetic effect with ufd-2 (RNAi) (Supplemental Figure S2C), characteristic expansion of oocytes (Supplemental Figure S2D), and reduction of sperm in the gonads of young adult hermaphrodites (Supplemental Figure S2E), supporting our idea that the C terminus of RPN-10 can cooperate with the UFD-2 pathway to metabolize the protein(s) required for production of male zygotes. These findings indicate that the C-terminal domain of RPN-10, which contains two sets of UIM, is essential for regulation of sex determination under the condition of ufd-2 reduction. In addition, we found that the ability of reproduction in rpn-10 (tm1180)/rpn-10 (tm1349) worms is identical to that of rpn-10 (tm1180) homozygote hermaphrodites.
RPN-10 Regulates the TRA-2–mediated Sex Determination Pathway
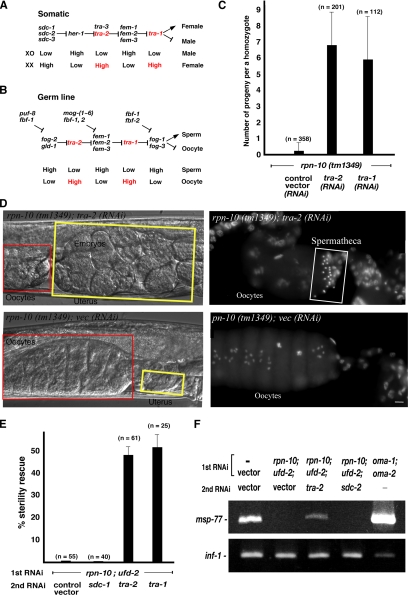
Next, we focused on the role of RPN-10 in specification of sexual fate. Most of the sexual fate decision genes have been identified and shown to function in a cascade of negative regulation (Ellis and Schedl, 2006; Zarkower, 2006). Figure 6A shows a somatic sex determination pathway that highlights functional relationships among genes at the end of the pathway. In hermaphrodite somatic cells, three SDC proteins inactivate the extracellular ligand HER-1, which blocks its transmembrane receptor protein TRA-2 (Meyer, 2000) (Figure 6A), and the pathway ultimately controls activity of the nuclear transcriptional factor TRA-1 (Zarkower, 2006). Figure 6B shows a simplified version of the germline sex determination (Goodwin and Ellis, 2002; Bachorik and Kimble 2005; Ellis and Schedl, 2006). In this pathway, the onset of hermaphrodite spermatogenesis depends on the control of TRA-2 germ-line activity. tra-1 is not a terminal sex regulator in germline, but the activity of tra-1 and tra-2 inhibits sperm production. Thus, both TRA-1 and TRA-2 promote feminization in the sex-determining pathways. We addressed the question of whether the defects in rpn-10 (tm1349) homozygote or rpn-10;ufd-2 (RNAi) worms would result in excess activation of the above proteins, which may lead to induction of inappropriate feminization (Figure 6, A and B). The results showed that tra-1 (RNAi) (which inhibits tra-1 expression to almost 55%) reversed the sterile phenotype of rpn-10 (tm1349) homozygote, supporting the view that TRA-1 and/or its upstream cascade pathway are responsible for the sterility of rpn-10 (tm1349) (Figure 6C). Knockdown of tra-2 by RNAi to >90% resulted in masculinization as reported previously in tra-2 mutants (Hodgkin and Brenner, 1977), but we found that partial knockdown of the tra-2 gene product (57% reduction of its mRNA) effectively rescued the sterile phenotype of rpn-10 (tm1349) homozygote (Figure 6C). The results of morphological observations supported the above results; knockdown of tra-2 with rpn-10 (tm1349) restored sperm formation and resulting viable embryos (Figure 6D). We obtained similar results using co-knockdown of rpn-10;ufd-2 (RNAi) (as knockdown at P0 generation) with either tra-1 or tra-2 (as a knockdown at F1 generation without affecting the efficacies of rpn-10;ufd-2 knockdown) (Figure 6E). The results of RT-PCR analysis indicated that coknockdown of tra-2 with rpn-10;ufd-2 restored the expression of msp-77, a sperm marker (Figure 6F). Considered together, the above-mentioned findings support the view that RPN-10 acts in the sex determination pathway at a point of tra-2 to regulate their activity negatively, either directly or indirectly, and thus promotes sperm production in the C. elegans hermaphrodite.
Figure 6.
Sterility induced by deletion of the rpn-10 gene was rescued by knockdown of tra-2. Schematic diagram of the somatic (A) and germline (B) sex determination pathway in C. elegans. Accumulation of TRA-2 and TRA-1 proteins, indicated by red, induces the feminization phenotype. (C) Results of a “rescue” experiment. RNAi of either tra-1 or tra-2 rescued the sterile phenotype of rpn-10 (tm1349) to produce normal viable F2 embryos. Control vector RNAi did not influence the sterile phenotype. (D) Oocyte maturation defects of rpn-10 (tm1349) can be restored by knockdown of the expression of tra-2 and tra-1. Note that the expanded oocytes and empty uterus observed in rpn-10 (tm1349) worms were restored in tra-2 (RNAi) individuals. Hoechst staining of dissected gonads from rpn-10 (tm1349);vector control (RNAi) and rpn-10 (tm1349);tra-2 (RNAi) worms. Restored spermatheca is indicated by a white box. Bar, 10 μm. (E) At P0 stage, all worms were treated with firstRNAi (rpn-10;ufd-2). This procedure ensures F1 sterility even in the absence of RNAi treatment at the F1 stage. From the young adult stage of P0 to F1 larval development, individuals were treated with second RNAi with a vector control, sdc-1 (RNAi), tra-1 (RNAi), and tra-2 (RNAi), respectively. The resulting number of F2 progeny per an F1 parent was counted. Second RNAi of tra-2 and tra-1 restored sperm formation. (F) RT-PCR analysis of the spermspecific marker msp-77. Expression of msp-77 is restored in rpn-10; ufd-2;tra-2 (RNAi) worms but not in rpn-10;ufd-2;vector control (RNAi) worms. inf-1 transcript amplified under similar conditions was used as a control.
RPN-10 Functions Cell Autonomously in the Germline
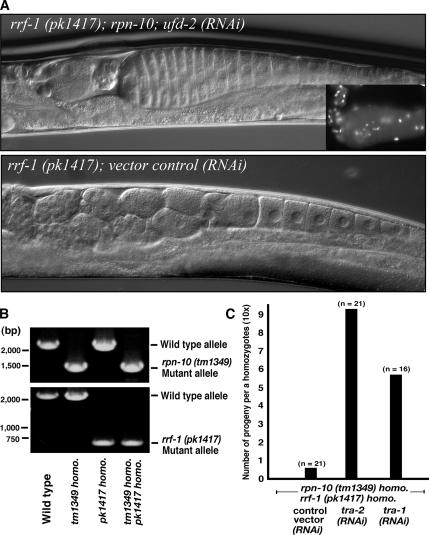
To verify whether the product of the rpn-10 gene acts cell-autonomously in the germline, we performed a feminization assay under an rrf-1 (pk1417) genetic background. In this mutant, RNA interference for genes expressed in somatic tissue was lost, whereas interference was retained for genes expressed in the germline (Sijen et al., 2001). We observed a typical sign of feminization of the germline in rrf-1 (pk1417) homozygotes that were treated with rpn-10; ufd-2 (RNAi) (Figure 7A), whereas control rrf-1 (pk1417) worms were viable, self-fertile, and none showed any obvious morphological or growth defects. These results support the notion that RPN-10 function in the germline is needed for the production of sperm. To show that the sex determination pathway is a target of RPN-10 in the germline, we established a double homozygote mutant of rrf-1 (pk1417);rpn-10 (tm1349) (Figure 7B). Although this double mutant worm shows less sterility to rpn-10 (tm1349) single homozygotes for an unknown reason, knockdown of either tra-1 or tra-2 in double mutants greatly enhanced self-fertility (Figure 7C). These results collectively suggest that the product of the rpn-10 gene functions cell autonomously in the germline tissues to promote male differentiation.
Figure 7.
RPN-10 acts cell-autonomously in the germline. (A) RNAi analysis under an rrf-1 (pk1417) genetic background, which is fully proficient for RNAi responses in the germline but not the soma. Nomarski observations indicate a typical sign of feminization of the germline in rrf-1 (pk1417) homozygotes that were treated with rpn-10; ufd-2 (RNAi). Inset, Hoechst staining of dissected gonads show absence of sperm in the spermatheca. Bar, 10 μm. (B) PCR verification of a double homozygote mutant, rpn-10 (tm1349); rrf-1 (pk1417). (C) Results of a “rescue” experiment. RNAi of either tra-1 or tra-2 rescued the sterile phenotype of rpn-10 (tm1349); rrf-1 (pk1417) to produce viable embryos. Control vector RNAi did not influence the sterile phenotype.
Knockdown of rpn-10 Overcomes the Germline-musculinizing Effect of Gain-of-Function Mutant of fem-3
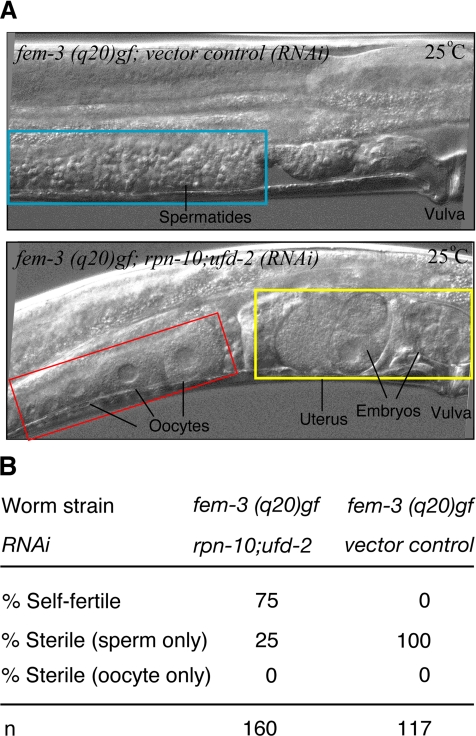
The fem genes are required for male development in the XO soma and for spermatogenesis in both XO and XX germlines. It has been reported that fem-3 (gf) alleles masculinize only hermaphrodite germlines (Barton et al., 1987). Thus, the XX fem-3 (gf) mutant at a restricted temperature (25°C) has a normal hermaphrodite soma, but the germline produces a vast excess of sperm and no oocytes. Interestingly, it has been reported that an intermediate allele of fem-3 (gf), q20, when homozygous in combination with tra-2 (gf) or fog-2 (lf), gave a fertile hermaphrodite at 25°C (Barton et al., 1987), suggesting that overproduction of tra-2 product during adulthood will counteract the excess FEM-3 produced by fem-3 (gf). We therefore investigated whether rpn-10;ufd-2 (RNAi) overcomes the masculinizing effect of fem-3 (gf) q20. We found that oocyte production was restored in >70% of fem-3 (gf) mutant worms by treatment with rpn-10;ufd-2 (RNAi) (Figure 8). The oocytes it makes can be fertilized and can give rise to progeny (Figure 8A). These results clearly indicate that, as has been reported for tra-2 (gf), co-knockdown of rpn-10 and ufd-2 pathway can suppress the masculinizing effect of fem-3 (gf).
Figure 8.
Co-knockdown of rpn-10 and ufd-2 overcomes the germline-masculinizing effect of fem-3 (gf). P0 larvae of fem-3 (gf) XX homozygous mutants were treated with either rpn-10;ufd-2 (RNAi) or control vector (RNAi) at a permissive temperature (20°C), and resulting F1 larvae were synchronized and continued to treated with respective RNAi at a restrictive temperature (25°C). Resulting F1 adult worms (55 h after hatching) were observed (A). In control vector (RNAi) worms, complete germline masculinization was observed, whereas >70% of rpn-10;ufd-2(RNAi)-treated F1 showed mutual suppression of germline-masculinizing phenotypes of fem-3 (gf), resulting in self-fertility at a restrictive temperature (25°C) (B).
Knockdown of rpn-10 Had No Obvious Effect on Morphology of XO Males but Caused Weak XO Intestinal Feminization
In XO males, secreted HER-1 is postulated to bind and to inactivate TRA-2A (Perry et al., 1993; Kuwabara, 1996). It has been reported that most gain-of-function alleles of tra-2 feminize the germline of XX animals but do not affect the germline or soma of XO males (Doniach, 1986; Schedl and Kimble, 1988). In overexpression of TRA-2 protein driven from a strong promoter, it has been reported that the germline of XO animals was transformed into hermaphrodites and partially feminized the intestine of XO animals (Kuwabara and Kimble, 1995). We therefore became interested in whether knockdown of rpn-10 could potentially feminize not only XX hermaphrodites but also XO males.
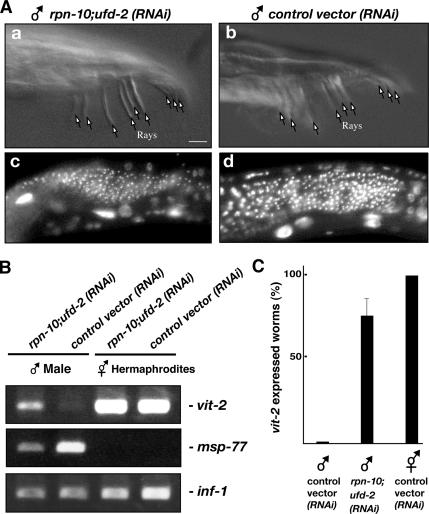
We found that simultaneous knockdown of rpn-10 and ufd-2 did not affect apparent morphology of XO males (Figure 9A) under the condition in which XX hermaphrodites were completely feminized. As shown in Figure 9A, tails of XO males treated with rpn-10;ufd-2 (RNAi) are undistinguishable from those of control males, and there is no sign of oocyte formation in the gonads. To detect the potential feminization in XO males more precisely, we performed semiquantitative RT-PCR analysis of somatic and germline sex determination markers. The C. elegans vitellogenin gene vit-2 is subjected to sex- and tissue-specific expression (Kimble and Sharrock, 1983). It is expressed solely in the adult hermaphrodite intestine, a female somatic tissue, and it has been reported that no vit-2 mRNA is present in the male or in any other cells of the hermaphrodite. We quantified the expression of vit-2 transcript in individual males to determine whether knockdown of rpn-10 influences the sexual fate of male somatic tissue. We found that vit-2 transcript is reproducibly up-regulated in XO males treated with rpn-10;ufd-2 (RNAi) (Figure 9, B and C), although the amount was less than that in genuine hermaphrodites. In contrast, the expression of msp-77, a sperm marker, is down-regulated in these XO males (Figure 9B). We could not detect significant up-regulation of oogenesis-specific transcripts such as rme-2 or mex-3. These results indicate that overall morphology of XO males is apparently not influenced by knockdown of rpn-10 but that it causes limited but reproducible XO intestinal feminization.
Figure 9.
Co-knockdown of rpn-10 and ufd-2 cause weak XO intestinal feminization. (A) Tail morphologies of rpn-10;ufd-2 (RNAi) and wild-type XO males (a and b). Hoechst staining of XO male gonads from rpn-10;ufd-2 (RNAi) and wild-type worms (c and d). Bar, 10 μm. (B) RT-PCR quantification of the somatic feminization marker vit-2 (422 base pairs, exons 4 and 5) and the sperm-specific marker msp-77 (360 base pairs) from a single individual worm. Note that the expression levels of msp-77 signal in hermaphrodites were too low to detect under the conditions used in this experiment. inf-1 transcript was used as a amplification control. (C) More than 70% individual males treated with rpn-10;ufd-2 (RNAi) expressed increased amounts of vit-2, whereas no wild-type male express it.
Defects in RPN-10 Result in Accumulation of TRA-2
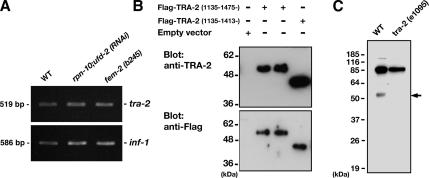
To test whether RPN-10 is involved in the control of TRA-2 accumulation, we examined TRA-2 in rpn-10–defective worms. Semiquantitative RT-PCR analysis showed that the steady-state levels of tra-2 transcript were similar in rpn-10;ufd-2 (RNAi) worms and N2 wild-type hermaphrodites (Figure 10A), indicating that the amount of tra-2 transcript is not influenced by rpn-10 knockdown. We then examined the accumulation of TRA-2 protein. The cellular behavior of endogenous TRA-2 protein is still enigmatic as a high titer antibody had not been available. To detect the endogenous TRA-2 ICD protein directly, we newly established an antiserum to the seventh intracellular domain of TRA-2 (TRA-2 ICD). As shown in Figure 10B, this antibody can specifically recognize TRA-2 ICD fragments expressed in mammalian cells and can detect antigens as efficiently as can high-performance anti-FLAG M2 antibody (Sigma-Aldrich). Furthermore, this antibody can detect proteins of 54 kDa as well as several lower-molecular-weight fragments in C. elegans extracts. These signals are specific to endogenous TRA-2 protein because tra-2 (e1095) putative null mutant worms show no such signals (Figure 10C). We speculate that the 54-kDa band corresponds to the endogenous TRA-2 ICD fragment (Sokol and Kuwabara, 2000). We currently have no idea about the physiological significance of smaller fragments.
Figure 10.
Establish of anti-TRA-2 ICD antibody. (A) RT-PCR-based expression analysis of tra-2 transcript (top; 519 base pairs, exons 22 and 23). inf-1 transcript (586 base pairs, exons 3–5) amplified under similar conditions was used as a control (bottom). (B) FLAG-tagged TRA-2 protein (fragments of amino acids 1135-1475 and 1135–1413, respectively) was expressed in COS7 cells, and the extracts were blotted with either an antibody against the C-terminal fragment (ICD) of TRA-2 or anti-FLAG M2 antibody (Sigma-Aldrich). (C) Fifty individual animals showing an appropriate phenotype were picked up and boiled with SDS sample buffer and then subjected to immunoblot experiments. The anti-TRA-2 ICD antibody can detect the endogenous 54-kDa band (indicated by an arrow) as well as several lower-molecular-weight fragments in the extracts. A tra-2 (e1095) putative null mutant was used as a negative control. Note that the 85-kDa signal is not specific because preimmune control serum stained the same band as well.
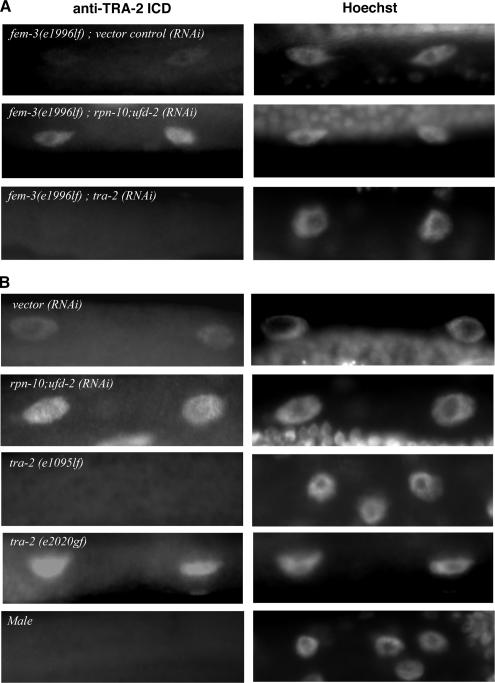
To examine the cellular behavior of TRA-2 ICD protein, we performed immunocytochemical experiments. We found that the nuclei of the hermaphrodite intestine, a female somatic tissue, were clearly immunostained with this anti-TRA-2 ICD antibody (Figure 11). The nuclear staining is specific to endogenous TRA-2 protein, as tra-2 (RNAi) on fem-3 (e1996lf) females greatly reduced the intensity of nuclear staining compared with that of the fem-3 (e1996lf) female itself (Figure 11A). Furthermore, there are essentially no nuclear signals in tra-2 (e1095lf) or wild-type males, whereas nuclei of tra-2 (e2020gf) were stained much stronger than that of wild-type (Figure 11B). For unknown reasons, immunosignals in the germline cells were difficult to detect under the experimental conditions we used. We speculate that the expression level of TRA-2 is much lower in the germline cells compared with that in intestinal cells. Thus, we currently have no immunological data of germline accumulation of TRA-2 protein.
Figure 11.
TRA-2 protein accumulates in RPN-10-defective worms. (A) fem-3 (e1996lf) is a putative null allele and developed as a true female regardless of whether tra-2 gene was knocked down or not. Even in a background of fem-3 (e1996lf), rpn-10;ufd-2 (RNAi) enhanced the nuclear accumulation of TRA-2, whereas tra-2 (RNAi) completely remove the staining. (B) The intestinal nuclei of rpn-10;ufd-2 (RNAi)-treated worms as well as gain-of-function mutant tra-2 (e2020) were stained more strongly than those of wild-type worms (vector RNAi control), whereas nuclei of loss-of-function mutant tra-2 (e1095) and wild-type male were scarcely immunostained with anti-TRA-2 ICD antibody. Note that all photographs were taken with identical exposure time. Bar, 10 μm.
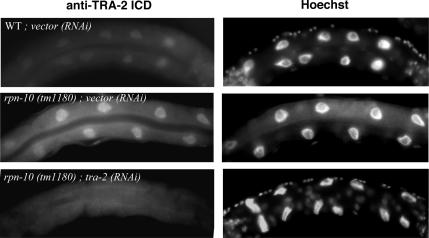
Interestingly, we found that rpn-10;ufd-2 (RNAi) treatment significantly increased the nuclear staining in the intestine (Figure 11, A and B). We determined the intensities of nuclear TRA-2 signals in fem-3 (e1996lf) females. The quantified results show that TRA-2 signal was strengthened for ∼2.8-fold in rpn-10;ufd-2 (RNAi);fem-3 (e1996lf) females compared with that in control females [fem-3 (e1996lf) with vector control (RNAi)]. These observations clearly support the notion that rpn-10 regulates TRA-2 protein levels irrespective of the sex differentiation backgrounds. We confirmed that the immunosignal of TRA-2 was completely absorbed by competition with excess corresponding antigens, and preimmune serum did not stain the nuclei of the intestine from wild-type hermaphrodite and rpn-10;ufd-2 (RNAi) females. Finally, we found that both rpn-10 (tm1180) and rpn-10 (tm1349) show significantly increased signal of TRA-2 in the nuclei of the intestine, and these nuclear stainings were eliminated by tra-2 (RNAi) (Figure 12). Lum et al. (2000) reported that an exogenous GFP-tagged fragment of TRA-2c, overproduced from the heat shock promoter, localized predominantly to nuclei in some somatic cells, and caused completely somatic feminization of XO animals. Our observations support the idea that endogenous TRA-2 protein was overaccumulated in the nuclei of feminized cells, presumably as ICD fragments, in rpn-10–defective conditions and thus induces excess feminization; forced reduction of tra-2 rescues this effect.
Figure 12.
rpn-10 mutant worm tm1180 shows significantly increased immunosignal of TRA-2 in nuclei of the intestine. The nuclear staining was eliminated by potent tra-2 (RNAi) treatment. Note that worms used in this experiment were fixed at the late larval stage (L4 to young adult). All photographs were taken with identical exposure time. Bar, 10 μm.
DISCUSSION
In the present study, we found that RPN-10, one of the proteasomal ubiquitin receptors, is required for appropriate sex determination in C. elegans. Knockdown of rpn-10 by RNAi sexually transforms F1 hermaphrodites to females by eliminating hermaphrodite spermatogenesis. The results of our analysis in rpn-10 (RNAi) C. elegans indicate that there is no detectable defect in the 26S proteasome architecture under normal growing conditions (Figure 2, D and E). In Drosophila, it was also reported that deletion of the subunit S5a/RPN-10/p54 does not disturb the assembly of the 26S proteasome (Szlanka et al., 2002). It has been reported that, in Drosophila, the deletion of the rpn10 gene caused larval–pupal lethality, multiple mitotic defects, and accumulation of polyubiquitinated proteins that were associated with increased amount of 26S proteasome subunits. In contrast to Drosophila, we did not detect obvious defects in the viability, life time, behavior, embryonic development, and stress tolerance in rpn-10 (RNAi)-treated P0 and F1 worms except for defects in F1 germ cells. Furthermore, knockdown of the expressions of any other proteasome subunits to any extent did not result in similar sterility. These observations suggest that the overall defects in the proteasome-mediated degradation pathway cannot explain the specific phenotype found in rpn-10 (RNAi) worms. We think that most of the ubiquitinated substrates were normally metabolized in rpn-10–defective worms by an alternative pathway for the polyubiquitinated substrate recognition system.
Normally, the hermaphrodite germline begins sperm formation in late L4 from a set of sperm precursor cells set aside in each half of the gonad. After a brief period of spermatogenesis, the germline of each half gonad switches from sperm to oocyte production. Based on the phenotype of gain-of-function alleles of tra-2, Doniach (1986) suggested that the activity of tra-2 is thought to be transiently repressed in the hermaphrodite germline (Doniach, 1986), thus allowing a burst of spermatogenesis to occur. In hermaphrodites, the TRA-2 activity is controlled by two posttranscriptional pathways. One post-transcriptional control of tra-2 activity is mediated through tra-2 3′ UTR binding proteins, such as GLD-1, which control tra-2 translation (Francis et al., 1995; Goodwin et al., 1997, Graves et al., 1999; Jan et al., 1999). The other is involved in regulation of TRA-2 activity by a posttranslational mechanism, identified as tra-2 (mx) (mixed character) mutations (Lum et al., 2000; Wang and Kimble, 2001). The regulation of TRA-2 pathway at the proteolytic level might allow further precise control of protein dosage. In this report, we suggest an additional layer of precise control over the activity of TRA-2 in XX hermaphrodites mediated by the RPN-10–dependent ubiquitin pathway.
Loss-of-function alleles of fog-2 and rpn-10;ufd-2 (RNAi) have similar feminization effects on the sexual phenotype of the hermaphrodite germline. The fact that both rpn-10;ufd-2 (RNAi), fog-2 (lf) and tra-2 (gf) suppress fem-3 (gf) to generate self-fertile hermaphrodites (Barton et al., 1987; Schedl and Kimble. 1988; this study) implies a common molecular pathway in these gene products. We also found that knockdown of rpn-10;ufd-2 results in potential somatic feminization in XO males. It was reported that overproduction of the intracellular domain of TRA-2 induced the expression of a vitellogenin reporter gene (vit-2::gfp) in transgenic XO animals (Lum et al., 2000). Although we found that vit-2 expression increased in rpn-10;ufd-2 (RNAi) XO animals, we also found that no increase in vit-2 expression was evident in tra-2 (e1095) null animals after rpn-10;ufd-2 (RNAi) treatment. Therefore, somatic feminizing activity induced by rpn-10 knockdown, although weak, was dependent on tra-2 activity. In addition, in XX hermaphrodite, the feminization phenotype of rpn-10 (tm1349) is rescued by knockdown of tra-2 or its downstream target tra-1, indicating that the TRA-2–mediated sex determination pathway is crucial for the rpn-10 (tm1349)-induced sterile phenotype. In accordance with this assumption, we detected the accumulation of TRA-2 by its specific antibody in rpn-10;ufd-2 (RNAi) worms as well as rpn-10–defective mutants. These observations are consistent with the view that the primary defect induced by RPN-10 removal is dysregulation in the TRA-2–mediated sex determination pathway; In larvae of hermaphrodite, timely inactivation of TRA-2 protein would lead to a lower TRA-2/FEM-3 ratio, resulting in transient spermatogenesis, whereas compromise of rpn-10 activity might abolish the appropriate down-regulation of TRA-2 and thus lead to feminization. Because of the intricacies of the germline sex determination pathway in C. elegans, the possibility that rpn-10 regulates another factor(s) in addition to TRA-2 remains. Among various known negative regulators of tra-2, fog-2 and rpn-10 defects have a phenotype similar to that of tra-2(gf) mutants: XX mutants are transformed into females, and XO mutants are apparently normal males (Schedl and Kimble, 1988; this study). In contrast, the other tra-2 repressor has distinct phenotypes; her-1 functions only in XO males (Trent et al., 1991). These findings indicate that the RPN-10–dependent ubiquitin pathway plays a role mainly in the onset of transient spermatogenesis to modulate sex-specific germ cell differentiation in hermaphrodites.
In yeast, it was recently proposed that there are at least two independent but closely related substrate delivery routes to the 26S proteasome: one route is the Rpn10p pathway that is more biased toward recognition of heavily ubiquitinated proteins, and the other route is the Ufd2p-Cdc48p-Rad23p(Dsk2p) route, which works through generation and recognition of proteins with a smaller number of ubiquitin molecules (Bazirgan and Hampton, 2005, Richly et al., 2005). Cdc48 seems to be particularly adapted to act on membrane proteins. For example, Cdc48 is crucial for the degradation of ER membrane proteins, because it seems to extract these proteins from the membrane after their ubiquitination and before their encounter with the proteasome (Elsasser and Finley, 2005). The lack of an overhanging structure, so-called Lid, may allow Cdc48 to unfold bulky ubiquitinatd proteins more readily, converting them into a state that is amenable for degradation by the proteasome. Many proteins that require Cdc48 to be degraded also require Ufd2 and Rad23 (or Dsk2, a Rad23 paralogue), suggesting that Rad23 links ubiquitinated targets of Ufd2 to the proteasome.
In this pathway, Rad23 binding to Ufd2 is critical for delivering targets from their site of ubiquitination to the proteasome. The transcription factor Spt23, many ERAD substrates, and the model substrate Ub-Pro-β-Gal all seem to be degraded by this pathway (Chen and Madura, 2002; Richly et al., 2005). In C. elegans, it has also been reported that UFD-2 and CHN-1 directly bind to UNC-45 and regulate UNC-45 protein level (Hoppe et al., 2004). In this study, we found that impairment of RPN-10 specifically induces F1 sterility, whereas knockdown of either ufd-2, rad-23, or dsk-2 or any combination of these genes did not, indicating for the first time the biological relevance of RPN-10 in C. elegans, and the RPN-10–mediated ubiquitin pathway is a major route for controlling sex determination. The fact that codepletion of ufd-2 with rpn-10 is required for the most penetrant effect suggests that UFD-2 (with RAD-23 and/or DSK-2) possesses a function partly overlapping the RPN-10–mediated route. It should be noted that even in the condition of rpn-10;ufd-2 (RNAi) (efficacy verified by RT-PCR and Western blot), there are no apparent defects in the P0 or F1 worm except for strictly restricted F1 reproductive abnormality. It could be explained that sex-determining pathway are highly sensitive to the partial retardation of ubiquitin–proteasome pathway, but this possibility seems unlikely as global and manifold abnormalities were induced by only a partial retardation of general 26S proteasome activity without preceded abnormality in reproduction (Figure 1 and Supplemental Tables SI and SII). We suggest the possible existence of alternative, as yet unidentified ubiquitinated substrate delivery route(s) to the 26S proteasome in C. elegans that is functionally distinct to the RPN-10–mediated ubiquitin recognition. This notion gives rise the interesting possibility that, in the ubiquitin-dependent pathway of higher eukaryotes, specificity of substrate discrimination is achieved not only by well-appreciated E3 ubiquitin–ligase diversity but also by an additional layer of the machineries that recruit the specific substrates to the proteasome by multiple ubiquitin receptors. It should be noted that knockdown of expression of RAD-23 and DSK-2, the other nonessential proteasomal ubiquitin receptors, did not induce reproductive defects, indicating that the induced sterility was specific to knockdown of rpn-10. These findings support the previously reported notion that the UFD pathway substrate Ub-Pro-βgal is remarkably stabilized in Rpn10p-deleted yeast, although the half-life of many short-lived proteins is unaffected (van Nocker et al., 1996; Fu et al., 1998). Interesting challenges for the future will be to determine the number of existing receptor pathways, sort out the mechanisms underlying the allocation of substrates to different receptor pathways, and determine whether individual receptor pathways are differentially regulated to modulate the repertoire of proteins degraded by the ubiquitin–proteasome system in response to specific cells. Accordingly, clarification of the substrate–recognition diversity by the RPN-10 family and other ubiquitin–recognition proteins is the future prospect.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. S. Mitani, K. Gengyo-Ando, T. Schedl, B. Grant, and the Caenorhabditis Genetics Center for providing the mutant strains and S. Harukuni for technical assistance. We also thank Drs. H. Sorimachi, Ken and Miyuki Sato, people in C. elegans society, and anonymous referees for many of constructive suggestions and discussions. M.S. is a recipient of research fellowship from the 21st Century COE Program. This work was supported in part by a grant from the Ministry of Education, Culture, Science, and Technology of Japan.
Footnotes
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-05-0437) on October 18, 2006.
REFERENCES
- Bachorik J. L., Kimble J. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc. Natl. Acad. Sci. USA. 2005;102:10893–10897. doi: 10.1073/pnas.0504593102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Schedl T. B., Kimble J. Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics. 1987;115:107–119. doi: 10.1093/genetics/115.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazirgan O. A., Hampton R. Y. Cdc48-Ufd2-Rad 23, the road less ubiquitinated? Nat. Cell Biol. 2005;7:207–209. doi: 10.1038/ncb0305-207. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R., Lee M.-H., Nayak S., Ohmachi M., Giordini F., Schedl T. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodites. Development. 2000;127:5265–5276. doi: 10.1242/dev.127.24.5265. [DOI] [PubMed] [Google Scholar]
- Chen L., Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detwiler M. R., Reuben M., Li X., Rogers E., Lin R. Two zinc finger proteins, OMA-1 and OMA-2, are redundantly required for oocyte maturation in C. elegans. Dev. Cell. 2001;1:187–199. doi: 10.1016/s1534-5807(01)00026-0. [DOI] [PubMed] [Google Scholar]
- Deveraux Q., Ustrell V., Pickart C., Rechsteiner M. A 26S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Doniach T. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics. 1986;114:53–76. doi: 10.1093/genetics/114.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R., Schedl T. Sex determination in the germ line (April 4, 2006) The C. elegans Research Community, WormBook. (WormBook, ed) 2006 doi: 10.1895/wormbook.1.82.2. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Elsasser S., Chandler-Militell D., Muller B., Hanna J., Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J. Biol. Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Elsasser S., Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Ferrell K., Deveraux Q., van Nocker S., Rechsteiner M. Molecular cloning and expression of a multiubiquitin chain binding subunit of the human 26S protease. FEBS Lett. 1996;381:143–148. doi: 10.1016/0014-5793(96)00101-9. [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Francis R., Maine E., Schedl T. Analysis of the multiple roles of gld-1 in C. elegans germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics. 1995;139:607–630. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Sadis S., Rubin D. M., Glickman M., van Nocker S., Finley D., Vierstra R. D. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J. Biol. Chem. 1998;273:1970–1981. doi: 10.1074/jbc.273.4.1970. [DOI] [PubMed] [Google Scholar]
- Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Goodwin E. B., Hofstra K., Hurney C. A., Mango S., Kimble J. A genetic pathway for regulation of tra-2 translation. Development. 1997;124:749–758. doi: 10.1242/dev.124.3.749. [DOI] [PubMed] [Google Scholar]
- Goodwin E. B., Ellis R. E. Turning clustering loops: sex determination in Caenorhabditis elegans. Curr. Biol. 2002;12:R111–R120. doi: 10.1016/s0960-9822(02)00675-9. [DOI] [PubMed] [Google Scholar]
- Graves L. E., Segal S., Goodwin E. B. TRA-1 regulates the cellular distribution of the tra-2 mRNA in C. elegans. Nature. 1999;399:802–805. doi: 10.1038/21682. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Falquet L. A. Ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. A., Brenner S. Mutations causing transformation of sexual phenotype in the nematode Caenorhabditis elegans. Genetics. 1977;86:275–287. [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., Albertson D. G. Isolation of dominant XO-feminizing mutations in Caenorhabditis elegans: new regulatory tra alleles and an X chromosome duplication with implications for primary sex determination. Genetics. 1995;141:527–542. doi: 10.1093/genetics/141.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe T., Cassata G., Barral J. M., Springer W., Hutagalung A. H., Epstein H. F., Baumeister R. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell. 2004;118:337–349. doi: 10.1016/j.cell.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Jager S., Schwartz H. T., Horvitz H. R., Conradt B. The Caenorhabditis elegans F-box protein SEL-10 promotes female development and may target FEM-1 and FEM-3 for degradation by the proteasome. Proc. Natl. Acad. Sci. USA. 2004;101:12549–12554. doi: 10.1073/pnas.0405087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E., Motzny C. K., Graves L. E., Goodwin E. B. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E. S., Ma P.C.M., Ota I. M., Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:1–10. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara H., et al. Developmentally regulated, alternative splicing of the Rpn10 gene generates multiple forms of 26S proteasomes. EMBO J. 2000;19:4144–4153. doi: 10.1093/emboj/19.15.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Sharrock W. J. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev. Biol. 1983;96:189–196. doi: 10.1016/0012-1606(83)90322-6. [DOI] [PubMed] [Google Scholar]
- Koegl M., Hoppe T., Schlenker S., Ulrich H. D., Mayer T. U., Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Kuwabara P. E. A novel regulatory mutation in the C. elegans sex determination gene tra-2 defines a candidate ligand/receptor interaction site. Development. 1996;122:2089–2098. doi: 10.1242/dev.122.7.2089. [DOI] [PubMed] [Google Scholar]
- Kuwabara P. E., Kimble J. Molecular genetics of sex determination in C. elegans. Trends Genet. 1992;8:164–168. doi: 10.1016/0168-9525(92)90218-s. [DOI] [PubMed] [Google Scholar]
- Kuwabara P. E., Kimble J. A predicted membrane protein, TRA-2A, directs hermaphrodite development in Caenorhabditis elegans. Development. 1995;121:2995–3004. doi: 10.1242/dev.121.9.2995. [DOI] [PubMed] [Google Scholar]
- Kuwabara P. E., Okkema P. G., Kimble J. Germ-line regulation of the Caenorhabditis elegans sex-determining gene tra-2. Dev. Biol. 1998;204:251–262. doi: 10.1006/dbio.1998.9062. [DOI] [PubMed] [Google Scholar]
- Kuwabara P. E., Perry M. D. It ain't over till it's ova: germline sex determination in C. elegans. Bioessays. 2001;23:596–604. doi: 10.1002/bies.1085. [DOI] [PubMed] [Google Scholar]
- Lum D. H., Kuwabara P. E., Zarkower D., Spence A. M. Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev. 2000;14:3153–3165. [PMC free article] [PubMed] [Google Scholar]
- Madura K. Rad23 and Rpn 10, perennial wallflowers join the melee. Trends Biochem. Sci. 2004;29:637–640. doi: 10.1016/j.tibs.2004.10.008. [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- Mehra A., Gaudet J., Heck L., Kuwabara P. E., Spence A. M. Negative regulation of male development in Caenorhabditis elegans by a protein-protein interaction between TRA-2A and FEM-3. Genes Dev. 1999;13:1453–1463. doi: 10.1101/gad.13.11.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B. J. Sex in the worm; counting and compensating X-chromosome dose. Trends Genet. 2000;16:247–253. doi: 10.1016/s0168-9525(00)02004-7. [DOI] [PubMed] [Google Scholar]
- Miller M. A., Nguyen V. Q., Lee M. H., Kosinski M., Schedl T., Caprioli R. M., Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Miller M. A., Ruest P. J., Kosinski M., Hanks S. K., Greenstein D. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 2003;17:187–200. doi: 10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M. D., Li W., Trent C., Robertson B., Fire A., Hageman J. M., Wood W. B. Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes Dev. 1993;7:216–228. doi: 10.1101/gad.7.2.216. [DOI] [PubMed] [Google Scholar]
- Pickart C. M. Targeting of substrates to the 26S proteasome. FASEB J. 1998;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- Pickart C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Puoti A., Pugnale P., Belfiore M., Schlappi A. C., Saudan Z. RNA and sex determination in Caenorhabditis elegans. EMBO Rep. 2001;2:899–904. doi: 10.1093/embo-reports/kve209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Saeki Y., Saitoh A., Toh-e A., Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 2002;293:986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- Schedl T., Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal S. P., Graves L. E., Verheyden J., Goodwin E. B. RNA-regulated TRA-1 nuclear export controls sexual fate. Dev. Cell. 2001;1:539–551. doi: 10.1016/s1534-5807(01)00068-5. [DOI] [PubMed] [Google Scholar]
- Shimada M., Kawahara H., Doi H. Novel family of CCCH-type zinc-finger proteins, MOE-1, -2, and -3, participates in C. elegans oocyte maturation. Genes Cells. 2002;7:933–947. doi: 10.1046/j.1365-2443.2002.00570.x. [DOI] [PubMed] [Google Scholar]
- Shimada M., Yokosawa H., Kawahara H. OMA-1 is a P granules-associated protein that is required for germline specification in C. elegans embryos. Genes Cells. 2006;11:383–396. doi: 10.1111/j.1365-2443.2006.00945.x. [DOI] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., Timmons L., Plasterk R.H.A., Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Sokol S. B., Kuwabara P. E. Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev. 2000;14:901–906. [PMC free article] [PubMed] [Google Scholar]
- Szlanka T., Haracska L., Kiss I., Deak P., Kurucz Z., Ando I., Viragh E., Udvardy A. Deletion of proteasomal subunit S5a/Rpn10/p54 causes lethality, multiple mitotic defects and overexpression of proteasomal genes in Drosophila melanogaster. J. Cell Sci. 2002;116:1023–1033. doi: 10.1242/jcs.00332. [DOI] [PubMed] [Google Scholar]
- Tanaka K. Molecular biology of proteasomes. Biochem. Biophys. Res. Commun. 1998;247:537–541. doi: 10.1006/bbrc.1998.8617. [DOI] [PubMed] [Google Scholar]
- Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Trent C., Purnell B., Gavinski S., Hageman J., Chamblin C., Wood W. B. Sex-specific transcriptional regulation of the C. elegans sex-determining gene her-1. Mech. Dev. 1991;34:43–55. doi: 10.1016/0925-4773(91)90090-s. [DOI] [PubMed] [Google Scholar]
- van Nocker S., Sadis S., Rubin D. M., Glickman M., Fu H., Coux O., Wefes I., Finley D., Vierstra R. D. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Oania R., Graumann J., Deshaies R. J. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wang S., Kimble J. The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO J. 2001;15:1363–1372. doi: 10.1093/emboj/20.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C. R., Seeger M., Hartmann-Petersen R., Stone M., Wallace M., Semple C., Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat. Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- Young P., Deveraux Q., Beal R. E., Pickart C. M., Rechsteiner M. Characterization of two polyubiquitin-binding sites in the 26S protease subunit 5a. J. Biol. Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- Zarkower D. Somatic sex determination (February 10, 2006) The C. elegans Research Community, WormBook. (WormBook, ed) 2006 doi: 10.1895/wormbook.1.84.1. 1.84.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.