Abstract
The dimerization of viral RNA through noncovalent interactions at their 5′ ends is a key step in the life cycle of retroviruses. In Moloney murine leukemia virus, three stem-loops are important in this process. One is a self-complementary tetraloop (H1), but the other two stem-loops (H2, H3) contain highly conserved GACG tetraloops that are not self-complementary sequences. Using two-dimensional NMR, we determined the structure of the H3 stem-loop. Surprisingly, it forms a stable, homodimeric kissing complex through only two intermolecular G⋅C base pairs. Cross-strand interactions of the adenines adjacent to the intermolecular G⋅C base pairs, plus unusual strong electrostatic interactions around the base pairs, contribute to the unexpected stability. This structure shows how even stem-loops without self-complementary sequences can facilitate the intermolecular recognition between two identical RNAs, and thus initiate dimerization and encapsidation of retroviral RNAs.
RNA dimerization is a key step in the life cycle of retroviruses (1–4) and is closely tied to the RNA encapsidation process (5–7). In Moloney murine leukemia virus, a dimer linkage structure overlaps the encapsidation domain (8, 9), where there are three conserved stem-loop structures (10) (Fig. 1A, H1, H2, and H3). Based on in vitro dimerization studies (11, 12), the H1 stem-loop was postulated to trigger RNA dimerization through base pairing of its self-complementary tetraloop. Two other stem-loop structures containing loop sequences that are not self-complementary tetraloops (H2, H3) also participate in this process (13). Because the H2 and H3 are essential for RNA packaging during encapsidation (9, 14), these two stem-loops may be key structural elements in connecting the dimerization and encapsidation processes. Both H2 and H3 hairpins contain GACG tetraloops that are highly conserved among murine type-C retroviruses (15) and that are thought to facilitate the recognition between the two genomic RNAs (13). However, it is not known how the H2 and H3 use their tetraloops in these processes. In this study, by using high-resolution NMR, the solution structure of the kissing complex of an 18-mer RNA oligonucleotide mimicking the H3 GACG tetraloop motif (H3–18; Fig. 1B) was determined. We found that GACG tetraloops form stable loop–loop kissing complexes.
Figure 1.
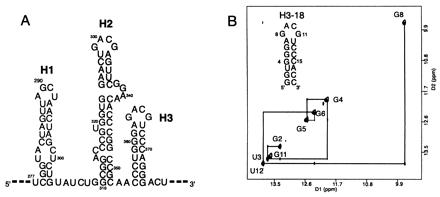
Characterization of H3 stem loop sequence. (A) The secondary structure of the H1, H2, and H3 stem loops of Moloney murine leukemia viral RNA (10). (B) The sequence and secondary structure of H3–18; note that the two terminal G⋅C base pairs are different from the native sequence. The imino proton region of the water NOESY spectrum (mixing time 300 ms, 283 K, 100 mM NaCl, pH 6.5) of H3–18 confirms the base pairing. The G8 and G11 imino protons were assigned based on the evidence described in the text.
Methods
RNA Sample Preparation and Characterization.
H3–18 RNA was transcribed from DNA templates in vitro by using T7 RNA polymerase (16). The preparation and purification of the RNA (17) and of the 13C, 15N-labeled NTPs (18) has been described. Native gel electrophoresis and RNA melting studies were done as described (17). The purified RNAs were heated at 95°C for 1 min and cooled to room temperature. For 15N-14N filtered nuclear Overhauser effect spectroscopy (NOESY) and 13C-12C heteronuclear multiple quantum coherence (HMQC)-NOESY-HMQC experiments, the same annealing procedures were done after mixing unlabeled RNAs with labeled RNAs.
NMR Spectroscopy.
NMR spectra were recorded on Bruker (Billerica, MA) DRX 500-MHz and Bruker AMX 600-MHz spectrometers. Spectra were processed by using felix 95.0 (Biosym Technologies, San Diego). One-dimensional (1D) jump-return experiments were done at various temperatures (Fig. 2). All of the two-dimensional (2D) NMR spectra were recorded in the same condition (100 mM NaCl/0.1 mM EDTA/10 mM sodium phosphate, pH 6.5). 2D NOESY, HNN-correlation spectroscopy (COSY) (19), 15N-1H heteronuclear single quantum coherence (HSQC), and 15N-14N filtered NOESY (20) spectra in 90% H2O/10% D2O were recorded at 10°C and 20°C. All other spectra were recorded in 99.96% D2O. Double quantum filtered (DQF)-COSY, phosphorus-decoupled high resolution DQF-COSY, homonuclear total correlation spectroscopy, proton-detected 31P-1H heteronuclear correlation spectroscopy HETCOR, and natural abundance 1H-13C HMQC (21) were recorded on the unlabeled H3–18. 2D 13C-12C HMQC-NOESY-HMQC (22) was done at 10°C and 20°C.
Figure 2.
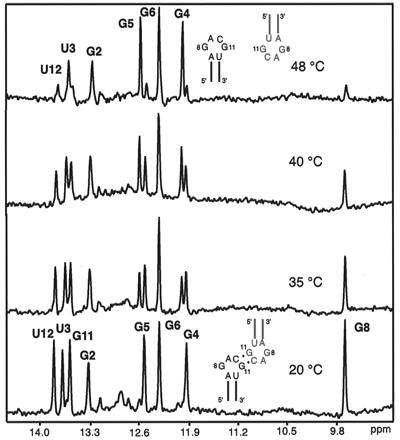
The imino spectra of H3–18 at different temperatures (0.1 mM RNA concentration, 50 mM NaCl, pH 6.5, 1,024 scans). The melting of the G8, G11, and U12 imino peaks occurs near the same temperature as the minor UV melting transition (48°C), whereas the G imino protons from the stem still show sharp peaks. The stem imino peaks G4 and G5 shift significantly with the transition from dimer to monomer, but G2, U3, and G6 do not. Schematics of a kissing dimer (20°C) and melted monomers (48°C) are shown.
NMR Assignments and Structural Calculation for H3–18 RNA.
All of the slowly exchanging imino and amino protons, base protons, and H1′ sugar protons were assigned by using H2O NOESY and D2O NOESY spectra, and were confirmed by 15N-1H HSQC and natural abundance 1H-13C HMQC. All of H2′, H3′, most of H4′, and a few H5′/H5′′ protons were assigned by using DQF-COSY, TOCSY, proton-detected 31P-1H HETCOR, and high-resolution DQF-COSY. A total of 528 distance constraints were used for structural calculations. Because the two molecules in the dimer seem to experience identical local magnetic environments evidenced by the presence of one set of cross peaks in H2O NOESY and D2O NOESY, the same NOE constraints were used for both molecules. Using D2O NOESY at different mixing times (50 ms, 100 ms, 150 ms, 200 ms, and 400 ms), distance constraints were classified into four categories, 1.8–3.2 Å, 2.0–4.0 Å, 2.5–5.0 Å, and 3.0–7.0 Å. Dihedral angle constraints for torsion angles δ, χ, and ɛ were obtained from analysis of DQF-COSY, D2O NOESY, and 31P-1H HETCOR. The ribose rings were constrained to 3′ endo conformations for all residues except for C10, G1, and C18 that show intermediate puckering between 2′ endo and 3′ endo. A-form geometry constraints were assumed for torsion angles α, β, γ, and ζ in the stems except for the closing base pairs. Based on 31P chemical shifts, α and ζ were loosely restrained to exclude a trans conformation for C10. The structure was calculated by using the x-plor 3.1 package (23) with restrained molecular dynamics (rMD). No symmetry constraints were used. Three consecutive protocols were used for the structural calculations: global fold, refinement, and final minimization (24). Fifty starting structures were generated with randomized torsion angles and were subjected to the initial round of global folding. Nineteen lowest energy structures were then refined and energy-minimized using a rMD-simulated annealing protocol (25).
Results
UV melting experiments on H3–18 RNA in 10 mM sodium phosphate buffer, pH 6.5, 50 mM NaCl revealed two transitions: a major transition with a melting temperature (Tm) of 82°C, and a broad minor transition with a lower Tm. The concentration dependence of the Tm of these two transitions was measured over a 200-fold concentration range (5 μM to 1 mM) in 50 mM NaCl. The Tm of the major transition did not change at all, whereas the minor transition showed a marked increase of Tm with increasing RNA concentration. A similar behavior of Tm was also observed in 100 mM NaCl. The concentration dependence indicates that the minor transition comes from the melting of an intermolecular dimer. This interaction was surprisingly strong; the dimer Tm was close to 60°C at RNA concentrations above 0.5 mM in 100 mM NaCl. This is consistent with a native gel electrophoresis result in which H3–18 runs as a monomer at room temperature, but as a smeared dimer band at 4°C (data not shown).
To determine the structural characteristics of the intermolecular complex, NMR spectra of H3–18 were measured in conditions corresponding to the dimer (RNA concentration 100 μM or higher/100 mM NaCl/10 mM sodium phosphate buffer, pH 6.5). The spectra showed only one set of NOE cross peaks corresponded to a single species, thus indicating the formation of a symmetric dimer. Water NOESY spectra confirmed the base pairing in the stem region by displaying all of the expected imino peaks (Fig. 1B). Two G imino protons from the tetraloop also showed sharp resonances, which indicate that the protons were protected from fast exchange with the solvent. One of the G imino protons (G8 in Fig. 1B) has a cross peak with the U imino proton (U12) of the closing A⋅U base pair; its up-field chemical shift (9.7 ppm) implies that it is protected but not hydrogen-bonded. The other G imino peak (G11 in Fig. 1B) is in the range of a normal G⋅C base pair (13.6 ppm), but has no cross peaks with any other imino protons. This G imino proton shows cross peaks with the two C10 amino protons (hydrogen bonded and nonhydrogen bonded), which indicates the formation of a Watson–Crick G⋅C10 base pair. An HNN-COSY experiment (19) confirmed that the N1 of the G forms a hydrogen bond with N3 of C10. The temperature dependence of the imino spectra (Fig. 2) showed that melting of the tetraloop and the loop closing base pair imino peaks (G8, G11, and U12) occurs near the Tm of the minor transition in the UV melting study (≈48°C). Using the condition favoring the monomeric conformation (0.5 mM H3–18, 25°C, 5 mM sodium phosphate, pH 6.5, no additional salt), we confirmed that the G8, G11, and U12 imino peaks are absent because of fast exchange with solvent in the monomeric form of H3–18. Moreover, the stem imino peaks (G2, U3, G4, G5, and G6) of the monomer H3–18 remain sharp and correspond to the sharp imino peaks of the kissing complex (see the 48°C spectrum in Fig. 2). This suggests that the observed minor transition at 48°C in the UV melting experiment is because of the transition from a kissing dimer to two monomers. The chemical shifts of G4 and G5 imino protons are down-field shifted in the monomer, although G6 is not. This is presumably caused by local conformational differences between the dimer and the monomer. Overall, these indicate that the G⋅C base pair in the loop is intermolecular.
To prove the existence of a loop–loop intermolecular complex, and to rule out the possibility of a double-helix dimer, an 15N-14N filtered-NOESY experiment (20) was done on a 1:1 mixture of the 15N-labeled and unlabeled H3–18. The experiment was designed to see only the cross peaks between protons attached to 15N and protons attached to 14N. As shown in Fig. 3A, the cross peaks between G imino and C amino protons for the Watson–Crick G⋅C base pairs were observed only for the base pair in the loop, not for the base pairs in the stem region. Therefore, only the G⋅C base pair in the loop is intermolecular; it confirmed the formation of a loop–loop symmetric kissing dimer rather than a duplex RNA. 2D HMQC-NOESY-HMQC 13C-12C filtering experiments (22) designed in a manner analogous to the 15N-14N filtered-NOESY experiments showed medium intermolecular NOEs (distance constraint 2.0 Å - 4.0 Å) between A9 H2 and C13 H4′, H5′/5′′ further confirming a kissing dimer (Fig. 3B).
Figure 3.
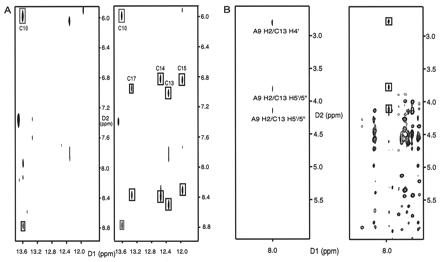
NMR evidence for the formation of the kissing complex of H3–18. (A) A comparison between the 15N-14N filtered-NOESY (Left) and the usual unfiltered water NOESY (Right). Both were done at 10°C (mixing time 300 ms, 100 mM NaCl, pH 6.5). The C13, C14, C15, and C17 cross peaks in the unfiltered water NOESY are between the two C amino protons and the G imino protons in the G⋅C base pairs for the stem. The C10 cross peaks are from the intermolecular G11⋅C10 base pair. Only the G11⋅C10 cross peaks are seen in the left spectrum. (B) A comparison between the 2D 13C-12C HMQC-NOESY-HMQC (Left) and the regular D2O NOESY (Right) in 100 mM NaCl, pH 6.5. Only cross peaks with black boxes in the right spectrum appear in the left spectrum showing the intermolecular NOEs; this indicates intermolecular interactions between A9H2 and C13 ribose protons.
The intermolecular G⋅C base pair in the loop was assigned to G11⋅C10 based on the following evidence. First, the regular D2O NOESY showed strong NOEs between H1′, H2′ of G8 and H5 of C10 that were confirmed as intramolecular NOEs by the absence of these NOEs in the 2D HMQC-NOESY-HMQC 13C-12C filtering experiments. This means that the G8 ribose is near the C10 base in the same molecule, which is not a possible geometry for the case of an intermolecular G8⋅C10 base pair. Second, if G8 formed an intermolecular base pair with C10, it would place an A9⋅A9 mismatch between the two G8⋅C10 base pairs. This is not compatible with the fact that A9 H2 is close to sugar protons of C13 of the other molecule shown by the presence of the medium intermolecular NOEs between them (Fig. 3B). Therefore, we can rule out G8⋅C10 and conclude that the intermolecular G⋅C base pair in the loop must be G11⋅C10.
Using 2D NMR experiments (see Methods), most of the base and sugar protons were assigned. In addition to the intermolecular NOEs described above, several weak or very weak NOEs between A9 H2 and G11 imino and H8 protons, U12 H1′, H2′, and C13 H3′, were also classified as intermolecular NOEs, because they would be close enough only in the context of the kissing dimer. In total, 32 intermolecular NOEs were used in the structure calculation of H3–18 (16 NOEs for each stem-loop).
Fifty random structures were started in the structural calculation protocol using x-plor (23). Nineteen structures converged with an rms deviation of 1.1 Å for the heavy atoms of the kissing loops including both loop closing A⋅U base pairs (Table 1). Fig. 4A shows the superposition of the 12 lowest energy structures. Because NOEs provide only short-range distance constraints (less than 7 Å), the angle between the two helices in the converged structures was variable (Fig. 4A). Few extremely kinked loop conformations were excluded by adding negative NOEs, based on the absence of the expected NOEs in the NMR spectra.
Table 1.
Structural determination statistics for the 19 converged structures of the H3-18 RNA kissing complex
| Number of NOE distance restraints | |
| Total number of the NOEs | 528 |
| Intramolecular-Intranucleotide NOEs | 182 |
| Intramolecular-Internucleotide NOEs | 298 |
| Intermolecular NOEs | 32 |
| Kissing loop with closing base pairs | 170 |
| Negative NOEs | 16 |
| Dihedral angle restraints (δ, χ and ɛ for all residues, α and ζ for C10) | 194 |
| A-form geometry dihedral restraints (α,β,γ,ζ) for the stems except for closing bps | 98 |
| Base planarity restraints | 32 |
| Total number of restraints | 852 |
| rms deviation for all heavy atoms relative to the average structure, Å | |
| The kissing loop with closing base pairs (A7–U12, A7′–U12′) | 1.1 ± 0.2 |
| Stem 1 (G1–G6, C13–C18) | 1.5 ± 0.5 |
| Stem 2 (G1′–G6′, C13′–C18′) | 1.2 ± 0.6 |
| NOE violations, Å | 0 (> 0.2 Å) |
| Angle violations, ° | 1 (> 5°) |
| Mean deviation from covalent geometry | |
| Bond length, Å | 0.004 |
| Angles, ° | 0.81 |
| Impropers, ° | 0.16 |
Figure 4.
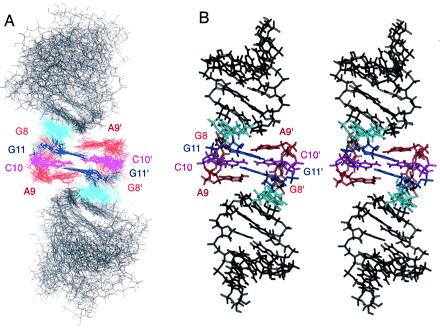
The structure of the H3–18 kissing complex. (A) The superposition of the 12 lowest energy structures, the curvature and the joining angle of the two stems are variable, although each stem shows reasonable rms deviation when superimposed separately (Table 1). The loop closing base pair A7⋅U12 is featured in sky-blue. (B) A stereoview of the averaged and energy-minimized structure of the H3–18 kissing complex.
The structural motif of the kissing loops contains two tandem intermolecular Watson–Crick G11⋅C10 base pairs capped by the A9 bases (Figs. 4B and 5A). This is quite different from the previously observed kissing dimers that contain six base pairs (26–28). The thermodynamic stability of this kissing complex was determined from the concentration dependence of Tm of the minor transition (29). The calculated standard enthalpy, ΔH°, was −29 kcal·mol−1, and the standard free energy at 37°C, ΔG°, was −6.5 kcal·mol−1 in 50 mM NaCl. This corresponds to the stability of four G⋅C base pairs (5′CGCG3′/3′GCGC5′) based on nearest neighbor parameters (29). The detailed structure shows the origin of this unusual stability. As seen in Fig. 5A, the two A9 bases partially stack on the tandem G11⋅C10 base pairs. Each adenine also has cross-strand interactions with the stem of the other molecule, as shown by the intermolecular NOEs between A9 H2 and sugar protons of U12 and C13. The two A9 base rings can act as barriers that protect the two intermolecular G11⋅C10 base pairs. The importance of A9 was supported by our mutational study showing that replacing A9 by U disrupts the dimer formation, as seen by native gel analysis. Purine bases in similar positions are also important in the kissing complex of SL1 in HIV-1 (28, 30).
Figure 5.
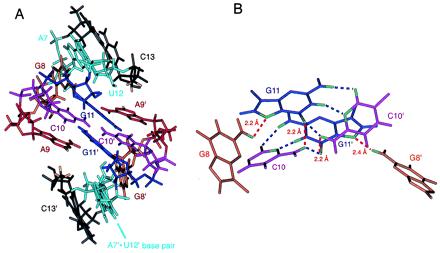
The structural characteristics of the H3–18 kissing complex. (A) A close view of the kissing loop region. The protons involved in the strong cross-strand interactions (A9 H2 and C13 H4′, H5′/5′′) are shown in yellow. (B) The network of hydrogen bonds and possible hydrogen bond-like electrostatic interactions around the intermolecular G⋅C base pairs. The blue lines represent the observed hydrogen bonds that are typical in Watson–Crick G⋅C base pairs, and the red lines represent the hydrogen bond-like strong electrostatic interactions.
Several unusual characteristics were observed for the G11⋅C10 base pairs. Most converged structures, including the energy-minimized average structure, show that the C10 amino protons are positioned not only in hydrogen bond distance and angle (less than 2.2 Å and 180° ± 60°) with the cross-strand keto group of G11 of the Watson–Crick base pair, but are also very close to the keto group of the same-strand G11 (Fig. 5B). These extra hydrogen bond-like electrostatic interactions around C10 amino protons might explain the down-field chemical shift of the hydrogen-bonded C10 amino proton (8.8 ppm) compared with the other hydrogen-bonded C amino protons. This unusual positioning of the C10 base is consistent with the observation that its H5-H6 NOE peak is weaker than others, and that C10 ribose has a mixture of 3′ endo and 2′ endo ribose conformations, implying that C10 is dynamic. All of the other residues have 3′ endo sugar conformations with the exception of the 5′ and 3′ end sugars. Fig. 5B also shows that one of the amino protons of G8 is near the N7 of G11, which is consistent with the NMR observation that the G8 amino protons are slowly exchanging with water and that they show well-separated chemical shifts (7.8 ppm, 7.2 ppm). Although the position of base G8 is not clearly visible in Fig. 5A, the structure shows that the G8/G8′ imino protons are well protected from solvent by neighboring nucleotides. This is consistent with the up-field chemical shifts of G8/G8′ imino protons described earlier.
Fig. 6 shows important stacking interactions around the kissing loops. The consecutive stacking of A9 bases sandwich the two G⋅C base pairs (Fig. 6 Left). There is considerable intermolecular stacking between the base of A9 and the ribose ring of C13 (Fig. 6 Middle), which accounts for the unusual up-field chemical shifts of H4′ (2.8 ppm) and H5′/H5′′ (3.8, 4.1 ppm) of C13. The bases on the ends of the tetraloop (G8, G11) stack on the closing A7⋅U12 base pair; G8 base stacks on A7 base and G11 base stacks on the ribose of U12 (Fig. 6 Right). This explains the U12 sugar H1′ up-field shift of 3.9 ppm. Because each G8 base stacks with the A7 base in the same molecule, the RNA strand undergoes an abrupt change in direction between G8 and A9 to close the loop. The break between G8 and A9 is consistent with about 1 ppm up-field shift of the A9 phosphorus compared with the chemical shifts of the stem phosphorus nuclei. Overall, these inter- and intramolecular stacking interactions and hydrogen bond-like strong interaction network around the two G⋅C base pairs contribute to the unusual stability of the kissing complex.
Figure 6.
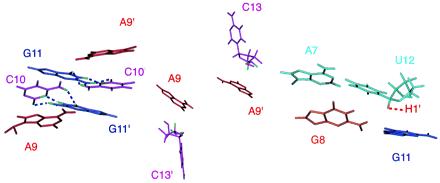
Important stacking interactions around the kissing loop. The stacking between the A9 base and the intermolecular G11⋅C10 base pair (Left). The hydrogen bonds in the G11⋅C10 base pairs are shown as blue dotted lines. The stacking between the A9 base and the C13 ribose with its H4′ and H5′/5′′ protons featured in green (Middle). The stacking between G8 base and the A7⋅U12 base pair, and the stacking between the G11 base and the U12 ribose with its H1′ in yellow (Right). The same stacking is observed in both hairpins.
Discussion
We found that the tetraloop sequence (5′GACG3′) forms an extra-stable kissing complex, although the sequence is not self-complementary. This may explain how the stem-loops H2 and H3 facilitate the dimerization and encapsidation processes of the viral RNA in Moloney murine leukemia virus (13). We speculate that strong kissing contacts between these stem-loops in two identical RNAs (H2⋅H2, H3⋅H3, or H2⋅H3) can be used as nucleation sites that allow other critical regions such as H1 to interact to complete the dimerization. This explains why the presence of H2 and H3 makes the dimerization process about seven times faster than the dimerization in their absence (13). Kissing complexes are well known to speed up base pair formation in naturally occurring antisense mechanisms (31).
There are changes in the secondary structure and the tertiary folding of RNA during the dimerization process (10); further changes may occur when the dimerized RNAs are packaged in the encapsidation process. The formation of the kissing complex through the conserved GACG tetraloops may be anchoring points that help initiate the conformational rearrangement of the RNA and facilitate its interaction with nucleocapsid proteins (6, 12). This study demonstrates a two base-paired RNA kissing complex that provides insight on the mechanism of the dimerization and the encapsidation of retroviral RNAs. It illustrates once again the surprises that RNA can provide.
Acknowledgments
We thank B. Dengler for general lab management and D. Koh for DNA template synthesis. We also thank J. Pelton for NMR advice and S.-H. Song and B.-Y. Hwang for their work in RNA sample preparation. Several colleagues read the manuscript and made helpful suggestions: R. Gonzalez, S. Lynch, J. Pelton, and J. Puglisi; we value their advice. This research was supported in part by National Institutes of Health Grant GM 10840, by the Department of Energy Grant DE-FG03-86ER60406, and through instrumentation grants from the Department of Energy (DE-FG05-86ER75281) and from the National Science Foundation (DMB 86-09305).
Abbreviations
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- HMQC
heteronuclear multiple quantum correlation
- HSQC
heteronuclear single quantum coherence
- 1D
one-dimensional
- 2D
two-dimensional
- COSY
correlation spectroscopy
- DQF
double quantum filtered
- rMD
restrained molecular dynamics
- Tm
melting temperature
Footnotes
Data deposition: The atomic coordinates of the H3–18 kissing complex have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1F5U).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170283697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170283697
References
- 1.Bender W, Davidson N. Cell. 1976;7:595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- 2.Bender W, Chien Y H, Chattopadhyay S, Vogt P K, Gardner M B, Davidson N. J Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paillart J C, Berthoux L, Ottmann M, Darlix J L, Marquet R, Ehresmann B, Ehresmann C. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clever J L, Parslow T G. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieth E, Gabus C, Darlix J-L. Nucleic Acids Res. 1990;18:119–127. doi: 10.1093/nar/18.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prats A C, Roy C, Wang P A, Erard M, Housset V, Gabus C, Paoletti C, Darlix J L. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout B, van Wamel J L B. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mann R, Mulligan R C, Baltimore D. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 9.Mougel M, Barklis E. J Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tounekti N, Mougel M, Roy C, Marquet R, Darlix J L, Paoletti J, Ehresmann B, Ehresmann C. J Mol Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]
- 11.Girard P M, Bonnet-Mathoniere B, Muriaux D, Paoletti J. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- 12.Girard P M, de Rocquigny H, Roques B P, Paoletti J. Biochemistry. 1996;35:8705–8714. doi: 10.1021/bi952454s. [DOI] [PubMed] [Google Scholar]
- 13.De Tapia M, Metzler V, Mougel M, Ehresmann B, Ehresmann C. Biochemistry. 1998;37:6077–6085. doi: 10.1021/bi9800303. [DOI] [PubMed] [Google Scholar]
- 14.Mougel M, Zhang Y, Barklis E. J Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konings D A M, Nash M A, Maizel J V, Arlinghaus R B. J Virol. 1992;66:632–640. doi: 10.1128/jvi.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milligan J F, Uhlenbeck O C. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 17.Sivakumaran K, Kim C-H, Tayon R, Jr, Kao C C. J Mol Biol. 1999;294:667–682. doi: 10.1006/jmbi.1999.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batey R T, Inada M, Kujawinski E, Puglisi J D, Williamson J R. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dingley A J, Grzesiek S. J Am Chem Soc. 1998;120:8293–8297. [Google Scholar]
- 20.Aboul-ela F, Nikonowicz E P, Pardi A. FEBS Lett. 1994;347:261–264. doi: 10.1016/0014-5793(94)00564-8. [DOI] [PubMed] [Google Scholar]
- 21.Varani G, Tinoco I., Jr J Am Chem Soc. 1991;113:9349–9354. [Google Scholar]
- 22.Folkers P J, Folmer R H A, Konings N H, Hilbers C W. J Am Chem Soc. 1993;115:3798–3799. [Google Scholar]
- 23.Brünger A T. x-plor: A System for X-ray Crystallography and NMR. New Haven: Yale Univ. Press; 1993. , Version 3.1. [Google Scholar]
- 24.Wimberly B T. Ph.D. thesis. Berkeley: University of California; 1992. [Google Scholar]
- 25.Varani G, Aboul-ela F, Allain F H-T. Prog NMR Spectrosc. 1996;29:51–127. [Google Scholar]
- 26.Marino J P, Gregorian R S, Jr, Csankovszki G, Crothers D M. Science. 1995;268:1448–1454. doi: 10.1126/science.7539549. [DOI] [PubMed] [Google Scholar]
- 27.Chang K-Y, Tinoco I. J Mol Biol. 1997;269:52–66. doi: 10.1006/jmbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- 28.Mujeeb A, Jared L C, Todd M B, James T L, Parslow T G. Nat Struct Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 29.Turner D H, Sugimoto N, Freier S M. Annu Rev Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- 30.Palliart J-C, Westhof E, Ehresmann C, Ehresmann B, Marquet R. J Mol Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 31.Eguchi Y, Itoh T, Tomizawa J. Annu Rev Biochem. 1991;60:631–652. doi: 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]


