Abstract
The adaptation mechanism of Pseudomonas aeruginosa ATCC 10145 to quaternary ammonium compounds (QACs) was investigated. A P. aeruginosa strain with adapted resistance to QACs was developed by a standard broth dilution method. It was revealed that P. aeruginosa exhibited remarkable resistance to N-dodecylpyridinium iodide (P-12), whose structure is similar to that of a common disinfectant, cetylpyridinium chloride. Adapted resistance to benzalkonium chloride (BAC), which is commonly used as a disinfectant, was also observed in P. aeruginosa. Moreover, the P-12-resistant strain exhibited cross-resistance to BAC. Analysis of the outer membrane protein of the P-12-resistant strain by two-dimensional polyacrylamide gel electrophoresis showed a significant increase in the level of expression of a protein (named OprR) whose molecular mass was approximately 26 kDa. The actual function of OprR is not yet clear; however, OprR was expected to be an outer membrane-associated protein with homology to lipoproteins of other bacterial species, according to a search of the National Center for Biotechnology Information website with the BLAST program by use of the N-terminal sequence of OprR. A correlation between the level of expression of OprR and the level of resistance of P. aeruginosa to QACs was observed by using a PA2800 gene knockout mutant derived from the P-12-resistant strain. The knockout mutant recovered susceptibility not only to P-12 but also to BAC. These results suggested that OprR significantly participated in the adaptation of P. aeruginosa to QACs, such as P-12 and BAC.
In recent years, disinfectants have been used indiscriminately because of our increased desire for cleanliness. The boom in “antibacterial” products has accelerated these trends. Furthermore, the indiscriminate use of disinfectants provides a hotbed for the adaptation of bacteria to those products and increases the spread of resistant bacteria. Similarly, as with many other disinfectants, bacterial resistance to quaternary ammonium compounds (QACs) has recently caused serious problems. Pseudomonas aeruginosa isolates have frequently been found to be resistant to QACs (3, 4, 5, 9, 11, 19, 27, 28). P. aeruginosa is ubiquitous in the environment and shows intrinsic resistance to high levels of QACs (18). The intrinsic resistance of this organism seems to be due to the cell wall and the cell membrane; for example, antimicrobials cannot easily access their sites of action because of the lower level of permeability of the outer membrane (14), and efflux pumps cause enhanced levels of efflux of antimicrobials (8). Moreover, since the species has a broad range of growth temperatures and possesses sturdy metabolism pathways, in addition to high levels of resistance to antibiotics and disinfectants, it is difficult to control with antimicrobials and has been shown to be an opportunistic pathogen and an infectious pathogen in hospitals. The phenotypic changes in P. aeruginosa cells corresponding to the increased levels of resistance to QACs have been investigated and reported. These include changes in the profiles of the outer membrane proteins (9, 28), fatty acids (3, 4, 5, 9, 11), lipids (9, 19), lipopolysaccharide (28), cell wall (5), and cell surface hydrophobicity (5, 9, 26) and changes in drug uptake and the zeta potential of the bacterial cell surface (9). However, it is still unclear how molecular mechanisms contribute to these phenotypic changes in the QAC-resistant P. aeruginosa strains. There are several reasons why it is difficult to reveal in detail the mechanisms of bacterial adaptation to QACs; for example, multiple mechanisms often participate in the adaptation of bacteria to disinfectants (16), and the bacterial phenotypic changes induced by the adaptation to QACs are frequently unstable (5, 11, 27). These characteristics make the investigation of bacterial adaptation more difficult.
We previously reported that P. aeruginosa isolates resistant to N-dodecylpyridinium iodide (P-12), a member of the QACs which is structurally similar to cetylpyridinium chloride, could successfully be obtained by a standard broth dilution method (26). The MIC of P-12 for the resistant strain was about 7.8 times higher than that for the wild strain, and the adapted resistance to P-12 was comparatively stable. In the present study, the changes in the outer membrane protein profiles between the wild strain and the P-12-resistant strain were compared in order to elucidate the mechanism of adaptation of P. aeruginosa to P-12. Because the adaptation of P. aeruginosa to P-12 was expected to occur due to changes in the components in the outer membrane, we thought it to be the first target or barrier to QACs. As a result, we have specified a new outer membrane protein whose level of expression was increased in the P-12-resistant strain. We also investigated the correlation between the expression of the protein and the adaptation of P. aeruginosa to QACs using gene knockout techniques.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are shown in Table 1. The bacterial strains were mainly grown in Luria-Bertani (LB) broth (1.0% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, and 0.5% [wt/vol] NaCl [pH 7.0]) or nutrient broth (Becton Dickinson, Sparks, Md.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| ATCC 10145 | Prototroph | |
| RP12 | Strain adapted to P-12 by adaptation treatment for 11 cycles | 26 |
| RP12K | PA2800 gene-knockout mutant derived from RP12 | This study |
| RBAC | Strain adapted to BAC by adaptation treatment for 11 cycles | This study |
| E. coli | ||
| JM109 | recA1 endA1 gylA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+laclqlacZΔM15] | 31 |
| BL21 (DE3) | F′ ompT hsdSB gal(λcI857 ind1 Sam7 nin5 lacUV5-T7gene1) dcm(DE3) | 2 |
| S17-1 | thi pro hsdR recA chr::RP-4-2 | 23 |
| Plasmids | ||
| pUC18 | Apr, a high-copy-number cloning vector | 31 |
| pGEX-2T | Apr, a GST-fusion protein expression vector | 24 |
| pOR-GST | Apr, a pGEX-2T derivative carrying the PA2800 gene between the BamHI site and the EcoRI site | This study |
| pGEM-T | Apr, a TA cloning vector | 15 |
| pACΩGm | Gmr Tetr, a donor vector of Gmr cassette carrying the Ω sequences | 22 |
| pOR-K01 | Apr, a pGEM-T derivative carrying the PA2800 gene amplified by Taq DNA polymerase | This study |
| pOR-K02 | Apr Gmr, a pOR-K01 derivative carrying the Gmr determinant amplified by Pfu DNA polymerase | This study |
| pMOB3 | A plasmid carrying a 5.8-kbp NotI fragment encoding sacB, Cmr, and oriT | 21 |
| pOR-K03 | Apr Gmr Sacr, a pOR-K02 derivative carrying the 5.8-kbp NotI MOB cassette from pMOB3 | This study |
Ap, ampicillin; Gm, gentamicin; Km, kanamycin; Cm, chloramphenicol; Sac, sucrose; Tet, tetracycline.
Chemicals.
P-12 was synthesized in our laboratory, as described previously (6). Benzalkonium chloride (BAC) was purchased from Takeda Pharmaceutical Co., Ltd. (Tokyo, Japan). The other chemicals used in this study were of commercially available reagent grade and were used without further purification.
Measurement of MICs.
The MICs of the various disinfectants were measured by a standard broth dilution method (10). The disinfectant solutions were diluted stepwise with fresh nutrient broth to the prescribed concentrations. Briefly, a 1.25-fold disinfectant dilution series was made by adding 4 ml of the disinfectant solution to 1 ml of nutrient broth. A culture of the bacteria to be tested was preincubated in LB broth for 18 h at 37°C and was diluted to a concentration of approximately 1.0 × 106 cells/ml with nutrient broth. A 0.5-ml portion of the prepared cell suspension was added to an equal volume (0.5 ml) of each of the dilution series, and the mixtures were then incubated at 37°C for 24 or 48 h. The MICs of the disinfectants for the bacteria was determined by visual inspection. The measurements were carried out three times, and the data are presented as the averages of the three measurements.
Evaluation of bactericidal effects of QACs.
The culture was used at the midexponential phase of growth. Cells were harvested (centrifugation at 2,500 × g for 15 min at room temperature) and resuspended in 10 ml of physiological saline. The QACs tested were dissolved to the prescribed concentrations in 5 ml of physiological saline. After each 50-μl aliquot of the cell suspension was mixed with the QAC solution, the mixture was incubated in a water bath at 37°C for 5 min with shaking. One-tenth volume of the mixture was then added to 5 ml of physiological saline containing 0.7% (wt/vol) Tween 80 and was left for 5 min without shaking to inactivate the QACs. The number of living cells in the mixture was determined by counting the numbers of CFU on LB agar plates.
Development of P. aeruginosa with adapted resistance to QACs.
The P. aeruginosa ATCC 10145 strain with adapted resistance to the QACs was developed by a previously reported standard broth dilution method (26). Briefly, a 1.25-fold QAC dilution series was made by using the nutrient broth described above for measurement of the MICs. The culture of P. aeruginosa preincubated in LB broth for 18 h at 37°C was diluted to a concentration of 1.0 × 106cells/ml with nutrient broth. A 0.5-ml portion of the cell suspension was added to an equal volume (0.5 ml) of each of the QAC dilution series. After incubation for 24 h at 37°C, the MICs of the QACs for the bacteria tested were determined. The bacteria that had grown in the presence of the highest concentration of the dilution series (that is, in the presence of a concentration below the MIC) were adjusted to 1.0 × 106 cells/ml with nutrient broth, and then the bacteria were placed in the next dilution and the MICs of the QACs were determined. The adaptation was continued by repetition of this procedure and was completed when no increase in the MIC was observed in the last three or four cycles.
Analysis of outer membrane protein profile.
The outer membrane fraction was prepared by the method of Nakajima et al. (13). The outer membrane protein profile was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (7) and two-dimensional PAGE (2D-PAGE) in an electrophoresis apparatus (TEP-2; Shimadzu Corporation, Kyoto, Japan). The electrophoresed gels were stained with Coomassie brilliant blue R250. Sequencing of the amino acid sequence of the outer membrane protein extracted from the acrylamide gel fragments from 2D-PAGE was carried out by APRO Science (Tokushima, Japan).
Preparation of genomic DNA.
The genomic DNA was prepared from P. aeruginosa ATCC 10145 and a P-12-resistant strain (strain RP12) by ultracentrifugation on CsCl gradients (20).
Preparation of a specific antibody against the outer membrane protein encoded by the PA2800 gene.
The entire coding region of the outer membrane protein encoded by the PA2800 gene was amplified by PCR with a primer set (primers PA2800gst-Fw and PA2800gst-Bw) (Table 2) and the genomic DNA of RP12 as the template. We confirmed that the sequences of the PA2800 gene of PAO1, ATCC 10145, and the P-12-resistant strain were identical. Each primer possessed an additional 8-bp sequence cleavable by BamHI and EcoRI at the 5′ end, respectively. The 100-μl reaction mixture, which contained 1 U of Pfu DNA polymerase (Promega, Madison, Wis.), was treated for 1 min at 95°C, followed by 30 cycles of 1 min at 95°C, 0.5 min at 65°C, and 1 min at 73°C before a final heating at 73°C for 5 min. The amplicon obtained was double digested with BamHI and EcoRI and ligated into the pGEX-2T plasmid (Amersham Biosciences AB, Uppsala, Sweden) that had been double digested with the same restriction enzymes. Escherichia coli JM109 competent cells were transformed with a recombinant plasmid (pOR-GST), after confirmation that the nucleotide sequence had been inserted by use of a DNA sequencer (ALF-red; Amersham Biosciences).
TABLE 2.
Nucleotide sequences of oligonucleotide primers used in this study
| Gene | Primer | Sequence |
|---|---|---|
| PA2800 | PA2800-Fw | 5′-ATAGAGGGTGTTGTTGACGTGC-3′ |
| PA2800-Bw | 5′-ATCCACTGAAACGACGAAAGGC-3′ | |
| PA2800gst-Fw | 5′-GCGGATCCGCCAGTGAAGAAGAC CCTTGGGAAAGC-3′ | |
| PA2800gst-Bw | 5′-GCGAATTCTTAGAAGTCGTCCTCC ACCTCGCCG-3′ | |
| aacCI | GmR-Fw | 5′-TATCACCAGCTCACCGTCTTTCAT-3′ |
| GmR-Bw | 5′-ACATTCTTGCCCGCCTGATGAA-3′ |
For the expression of a fusion protein of glutathione S-transferase (GST) and the putative protein encoded by the PA2800 gene, E. coli JM109 transformed with pOR-GST was preincubated overnight in 15 ml of LB broth containing 50 μg of ampicillin per ml at 37°C. The preincubated culture (5 ml) was inoculated into 250 ml of LB broth containing 50 μg of ampicillin per ml, and the mixture was incubated at 37°C until the optical density at 600 nm of the culture reached 0.4 to 0.6; then, protein expression was induced by the addition of 1.0 mM (final concentration) isopropyl-thio-β-d-galactopyranoside. The induction of protein expression was performed at 37°C for 4 h, and the bacteria were harvested by centrifugation (1,700 × g for 10 min at 4°C). The bacterial cells were resuspended in 15 ml of phosphate-buffered saline and disrupted by three treatments with a French pressure cell (SLM Instruments, Inc., Urbana, Ill.) at 1,000 lb/in2. The disrupted sample was separated by SDS-PAGE, and the electrophoresed gel was stained with Coomassie brilliant blue R250. The overexpressed fusion protein consisting of GST and the putative protein encoded by the PA2800 gene were clipped off with a sterile scalpel. The fusion protein was extracted overnight in extraction buffer (20 mM Tris-HCl [pH 8.0] containing 1.0% [wt/vol] SDS) with shaking at room temperature. The extracted fusion protein was then precipitated with a fourfold volume of cold acetone. The fusion protein was obtained by centrifugation (20,000 × g for 30 min at 4°C). The identity of the extracted fusion protein was confirmed by 2D-PAGE, which gave a single spot.
A male Japanese White rabbit (weight, 2.0 kg) was immunized with the purified fusion protein consisting of GST and the putative protein encoded by the PA2800 gene. The fusion protein solution (100 μg of fusion protein in 1.5 ml of phosphate-buffered saline) was emulsified with an equal volume of Freund's complete adjuvant (Difco, Detroit, Mich.), and then the emulsion was subcutaneously administered to the back of the rabbit as a primary immunization. After 3 weeks, 3.0 ml of the emulsion of the fusion protein and Freund's incomplete adjuvant (Difco) was injected as a booster in the same manner as the primary immunization. The additional booster was administered two more times as described above. One week after the last injection, blood was taken from the ear vein to obtain the antiserum. The antiserum was stored at −30°C with 0.1% (wt/vol) sodium azide until use.
Immunoblotting.
Immunoblotting was carried out basically as described previously (12). Detection of the putative protein encoded by the PA2800 gene was performed with an antiserum specific for the putative protein encoded by the PA2800 gene as the first antibody, horseradish peroxidase-conjugated goat immunoglobulin G against rabbit immunoglobulin G (ICN Biomedicals, Inc., Aurora, Ohio) as the second antibody, and a chemoluminescence substrate kit (ECL Western blotting detection reagents; Amersham Biosciences).
Preparation of a PA2800 gene knockout mutant.
The preparation of a PA2800 gene knockout mutant was carried out by the conjugation method. The 1.0-kbp fragment containing the open reading frame of the PA2800 gene with its upstream and downstream sequences was amplified by using Taq DNA polymerase (Promega) and a primer set (primers PA2800-Fw and PA2800-Bw; Table 2). The mixture was treated for 5 min at 94°C, followed by 35 cycles of 1 min at 94°C, 1 min at 65°C, and 1 min at 72°C before the reaction was completed by treatment at 72°C for 10 min. The amplified fragment was subcloned into the pGEM-T vector (Promega) to yield pOR-K01. Subsequently, the 1.1-kbp fragment of the aacC1 gene encoding the gentamicin acetyltransferase-3-1 (Gmr) (22) was amplified by using the Pfu DNA polymerase (Promega), pACΩGm (22), and a primer set (primers GmR-Fw and GmR-Bw; Table 2). The mixture was treated for 1 min at 95°C, followed by 30 cycles of 1 min at 95°C, 0.5 min at 65°C, and 2 min at 73°C before a final treatment at 73°C for 5 min. The fragment was ligated into pOR-K01 at the BalI site in the middle of the PA2800 gene to yield pOR-K02. The MOB cassette carrying a chloramphenicol resistance (Cmr) determinant, oriT and sacB, was then excised from pMOB3 (21) as a 5.8-kbp NotI fragment. The fragment was ligated into the NotI site of pOR-K02 to obtain pOR-K03.
pOR-K03 was introduced into a mobilizer strain, E. coli S17-1, by electroporation. Conjugal transfer was achieved by the method of Schweizer et al. (21), with a slight modification. Early-exponential-phase cultures of both the donor strain (E. coli S17-1) and the recipient strain (P. aeruginosa RP12) were used. Both strains (100 μl of culture volume for each strain) were placed onto a sterilized filter (cellulose nitrate filter; pore size; 0.2 μm, diameter; 25 mm; Advantec, Tokyo, Japan) on an LB plate, and the plates were incubated overnight at 37°C. The cells on the filter were suspended in 2 ml of LB broth and centrifuged (6,500 × g for 5 min at 4°C). Aliquots of the pelleted cells were resuspended in 0.1 ml of LB broth and plated onto a NAC agar plate (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) to select only P. aeruginosa cells from the mixed culture of P. aeruginosa and E. coli. After incubation overnight at 37°C, the colonies that had grown were suspended in 10 ml of LB broth and the mixture was centrifuged (6,500 × g for 5 min at 4°C), and then the cells were resuspended in 1 ml of LB broth and a 0.1-ml portion was plated onto an LB plate containing 20 μg of gentamicin per ml to effectively induce gentamicin resistance. After an overnight incubation at 37°C, the colonies that had grown were plated onto LB plates containing 200 μg of gentamicin per ml and incubated as described above. The colonies that had grown were further plated onto LB plates containing 200 μg of gentamicin per ml and 5% (wt/vol) sucrose. After three rounds of selection on plates containing gentamicin and sucrose in order to ensure that recombination between the genomic DNA of P. aeruginosa RP12 and the plasmid DNA transferred from E. coli S17-1 had occurred, the PA2800 gene-knockout mutants were obtained as gentamicin- and sucrose-resistant colonies. Confirmation of the disruption of the PA2800 gene was checked by PCR amplification of the gene and immunoblotting of the bacterial proteins.
RESULTS AND DISCUSSION
P. aeruginosa with adapted resistance to QACs.
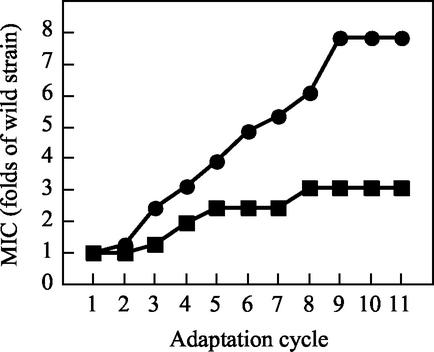
In a clinical or food production environment, it is thought that the concentration of disinfectants may be diluted to the sub-MIC for use in disinfection and rinsing. The exposure of bacterial cells to sub-MICs may give a selective pressure for resistance to the bacteria (29). In order to investigate the adaptation of P. aeruginosa to QACs under such conditions, repeated measurement of the MIC was adopted since the sub-MIC condition was easily estimated. Development of the adapted resistance of P. aeruginosa to the QACs is shown in Fig. 1. In the case of the adapted resistance of P. aeruginosa to P-12, the MIC constantly increased and reached the maximum of 384 ppm (7.8-fold higher than MIC for the wild strain). On the other hand, the adaptation of P. aeruginosa to BAC was weaker than that to P-12; nevertheless, the MIC constantly increased and reached the maximum of 51.2 ppm (3.0-fold higher than MIC for the wild strain). These results suggest that the adaptation of P. aeruginosa to the QACs (pyridinium-cationic-type QACs and ammonium-cationic-type QACs) was easily attained through continuous contact with the QACs. We mainly used the P-12-adapted strain (RP12) in the study. We did so because the molecular change(s) with adaptation was expected to be more remarkable in RP12 than in the BAC-adapted strain (strain RBAC), judging from the more significant increase in the MIC for RP12 than for RBAC. This adapted resistance to P-12 was comparatively stable in the strain (data not shown), but when the strain was kept at −30°C in the presence of 15% (vol/vol) glycerol, the resistance decreased slightly (see the MIC of P-12 for P. aeruginosa RP12 in Table 3). As shown in Table 3, RP12 had notable cross-resistance to BAC.
FIG. 1.
Relationship between adaptation cycle and MICs of QACs for P. aeruginosa ATCC 10145. Symbols: •, P-12; ▪, BAC. The adaptation of P. aeruginosa to the QACs was done as described in the Materials and Methods section. The adaptation was completed when no increase in the MIC was observed in the last three or four cycles.
TABLE 3.
MICs of P-12 and BAC for stationary-phase bacteria
MICs were measured by a broth dilution method with nutrient broth at 37°C for 24 h. The data are represented as the average values of three measurements.
A frozen stock (−30°C) culture recovered by incubation in LB broth at 37°C for 18 h was used.
Also, a gradual increase in the MIC of P-12 for P. aeruginosa was observed (Fig. 1), and this pattern of a gradual increase in the MIC was reproducible in separate experiments (data not shown). The MIC of P-12 for the frozen stock (−30°C) culture of RP12 decreased (Table 3), and the decreased MIC was recovered by contact with P-12 (data not shown). These facts suggest that the adapted P-12 resistance of P. aeruginosa was not the result of a mutation but the result of an adaptation. A similar explanation was seen for an isothiazolone-resistant P. aeruginosa strain (1, 30). The reason is that if P-12 adaptation was caused by a mutation, the decrease in the MIC can hardly be expected during preservation. However, the possibility of the participation of a mutation(s) in the resistance of P. aeruginosa to P-12 could not be completely excluded only on the basis of these results.
Analysis of change in outer membrane protein profile with adaptation.
It is well known that QACs such as P-12 and BAC can exclude boundary divalent cations such as Mg2+ from the outer membrane (28) and induce disorder in the outer and cell membranes. Therefore, the results observed here suggest that some change(s) that allowed the bacterium to resist the antibacterial action of the QACs occurred in the outer membrane, which is the first target or barrier of P. aeruginosa strains adapted to QACs, such as RP12 and RBAC. Therefore, in order to elucidate the adaptation mechanism of P. aeruginosa to the QACs, the change in the outer membrane protein profile was primarily investigated.
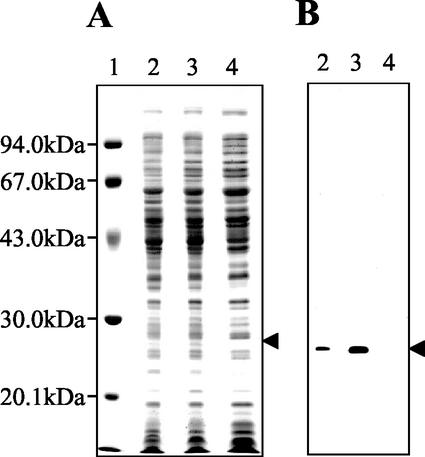
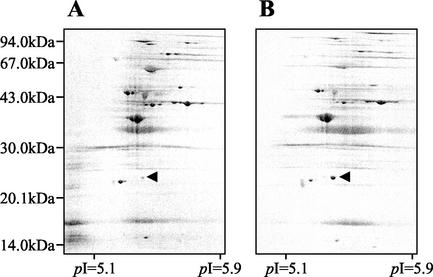
The outer membrane fractions prepared from P. aeruginosa ATCC 10145 and RP12 were analyzed by electrophoresis. Compared to the band pattern of P. aeruginosa ATCC 10145, some differences were seen in that of RP12 (Fig. 2A; compare lanes 2 and 3). In particular, the level of expression of the protein whose molecular mass was approximately 26 kDa was significantly increased in RP12. To investigate whether the 26-kDa protein band was composed of a single protein, 2D-PAGE was done and the pattern of the protein spots was observed. As shown in Fig. 3, the 26 kDa-protein was detected as a single spot; the increase in the level of expression of the 26-kDa protein was also confirmed in the 2D-PAGE image. Thus, it was obvious that the increase in the amount of the 26-kDa protein band shown in Fig. 2A, lane 3, reflected the increase in the amount of a single protein. These results indicate that the level of expression of the 26-kDa outer membrane protein was specifically increased in RP12. We named this 26-kDa protein OprR and further investigated the correlation between the QAC resistance of P. aeruginosa and the expression of OprR.
FIG. 2.
Electrophoretic analysis and immunoblotting of the strains tested. The arrowhead indicates OprR, whose expression was specifically increased in RP12 (approximate molecular mass, 26 kDa). (A) SDS-PAGE image of whole-cell extracts of P. aeruginosa ATCC 10145, RP12, and RP12K. Lanes: 1, molecular mass marker; 2, P. aeruginosa ATCC 10145; 3, RP12; 4, RP12K. (B) Immunoblotting of OprR. The contents of the lanes are the same as those in panel A. An antiserum against a fusion protein consisting of GST and the putative protein encoded by the PA2800 gene was used as the primary antibody.
FIG. 3.
2D-PAGE analysis of the outer membrane protein profiles of P. aeruginosa ATCC 10145 and RP12. (A) P. aeruginosa ATCC 10145; (B) RP12. The arrowhead indicates the OprR (approximate molecular mass, 26 kDa; pI = 5.3).
Molecular specification of OprR.
Amino acid sequencing of the N-terminal side of OprR was carried out, and the 12 residues were determined to be ASEEDPWESINR. A search of the website of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) was conducted with the BLAST program to find sequences homologous to the amino acid sequence. As a result, the putative product of an open reading frame, PA2800, whose function has been unclear, showed a complete match with the amino acid sequence of OprR. Judging from the actual amino acid sequence of OprR (ASEEDPWESINR), it was suggested that OprR was first translated as a proprotein and then processed into the mature form of OprR by removal of the N-terminal 30 amino acids, which encode a signal peptide. Also, by consideration of its primary structure, as deduced from the gene sequence, it is expected that OprR is a membrane-associated protein and would interact with QACs in the outer membrane because the deduced mature OprR contains hydrophobic regions (data not shown). Moreover, the antiserum against a fusion protein consisting of GST and this putative protein encoded by the PA2800 gene specifically recognized the 26-kDa protein whose levels were increased in RP12 (Fig. 2B, lane 3). Thus, it was concluded that OprR was encoded by the PA2800 open reading frame. The BLAST search also showed homology between OprR and the lipoproteins of several species, such as VacJ (Fig. 4). Although the vacJ gene of Shigella flexneri has been shown to be associated with the intercellular spread of the bacterium, it is still unclear whether the gene participates in resistance to QACs (25). If OprR is similar to the ordinary lipoproteins of the outer membrane, OprR may play a role in the mechanical enhancement of the resistance of the outer membrane to QACs. Alternatively, if OprR can associate with itself to form a channel in the outer membrane, this protein may act as part of a certain disinfectant efflux system. Moreover, if OprR functions as a member of some efflux pump system, the increase in the level of OprR would introduce an increase in the level of QAC efflux and would be consistent with the acquisition QAC resistance. However, no significant structural homology between the OprR and the components of the known efflux pump system has so far been found.
FIG. 4.
Multiple-sequence alignment of proteins homologous with OprR. The search for sequences homologous with OprR was performed with the BLAST program, and the multiple-sequence alignment was performed with CLUSTALX software. The conserved sequences are denoted in gray. Abbreviations: PA, P. aeruginosa; PP, Pseudomonas putida; PM, Pasteurella multocida; VC, Vibrio cholerae; EC, E. coli; SF, S. flexneri; SE, Salmonella enterica; RP, Rickettsia prowazekii; HI, Haemophilus influenzae.
Construction of a PA2800 gene-knockout mutant as an OprR deletion mutant.
To precisely investigate the effect of OprR expression in RP12, a PA2800 gene-knockout mutant was prepared from RP12. In order to confirm that the gene recombination occurred correctly on the genomic DNA of a PA2800 gene-knockout mutant, PCR amplification was done with a primer set which can amplify a sequence from about 1.0 kbp upstream of the PA2800 gene to about 1.0 kbp downstream of the gene. Since a single DNA band amplified from a PA2800 gene-knockout mutant was about 1.0 kbp larger than those from the wild strain and RP12, it was indicated that the original PA2800 gene in RP12 was replaced by the PA2800 gene into which a gentamicin resistance gene cassette from pOR-K03 was inserted (data not shown).
To confirm the lack of expression of OprR in a PA2800 gene-knockout mutant, the entire cell protein of a PA2800 gene-knockout mutant was electrophoresed and then detected by immunoblotting with antiserum against OprR as the primary antibody. As shown in Fig. 2B, lane 4, OprR was not expressed in the PA2800 gene-knockout mutant. As a consequence, we successfully constructed a PA2800 gene-knockout mutant of RP12 from which OprR was deleted. The knockout mutant obtained was named RP12K.
Characterization of a PA2800 gene-knockout mutant of P. aeruginosa.
Subsequently, we carried out a further investigation to evaluate the contribution of the expression of OprR to the adaptation of P. aeruginosa to the QACs by using RP12 and its PA2800 gene-knockout mutant, RP12K. As shown in Table 3, the MIC of P-12 for RP12K decreased significantly compared to that for the parent strain, RP12. This result indicates that RP12K became more susceptible to P-12 by the deletion of OprR. The MIC of BAC for RP12K was also investigated. Interestingly, the same trend was observed. The MIC of BAC for RP12K also decreased significantly compared to that for RP12. These results suggest that the increase in the level of OprR expression in RP12 significantly affects not only the adaptation to P-12 but also the adaptation to BAC. Therefore, we thought that OprR is one of the important proteins responsible for the adaptation of P. aeruginosa to the QACs. These results may also indicate that the resistance system involving OprR generally plays a key role in the resistance to the QACs. However, in order to confirm this hypothesis, the susceptibilities of RP12 and RP12K to various kinds of QACs must be extensively compared. We note that the extent of resistance of RP12K to P-12 and BAC was considerably lower than that of RP12, but RP12K was still obviously more resistant to both QACs than the wild strain, ATCC 10145, since the decreased MICs of both QACs for RP12K had not reached the same level as that for the wild strain. This tendency was also observed in the evaluation of the bactericidal effects of P-12 and BAC. The rate of survival of RP12K cells was much lower than that of RP12 cells, although RP12K cells still exhibited a higher survival rate than cells of the wild strain (data not shown). These results not only may suggest that the adaptation of P. aeruginosa to the QACs occurred as a result of the increase in the level of OprR expression, but they also may support the accepted idea that multiple factors contribute to the adaptation of the bacteria to disinfectants (16). Therefore, further investigation is necessary to elucidate the entire aspect of the adaptation of P. aeruginosa to QACs, e.g., changes in lipids or lipopolysaccharides.
The increase in the level of OprR expression observed in RP12 is thought to occur by a kind of stress response system. In gram-negative bacteria, it has been reported that multidrug resistance is activated by inductions and mutations caused by certain types of stresses (17). The bacterial response to environmental stress is an indispensable mechanism for survival under stressed conditions. Interestingly, PA2800 is located downstream of the genes encoding a hypothetical transcription regulator on P. aeruginosa PAO1 chromosomal DNA (http://www.pseudomonas.com/AnnotationByPAU.asp?PA=PA2802/). Therefore, this location of the PA2800 gene suggests that the regulation of OprR expression is controlled by stress, i.e., contact with QACs.
This study is the first step in the clarification of the molecular mechanism of the adaptation of P. aeruginosa to QACs; however, we have not obtained enough knowledge about this mechanism. In order to prevent the further expansion of QAC-resistant bacteria that can cause infections in hospitals and contaminate foods and cosmetics, elucidation of the molecular mechanism of resistance must be achieved soon. Further investigations are in progress to elucidate the mechanism of the adaptation of P. aeruginosa to QACs.
Acknowledgments
We are grateful to H. P. Schweizer, Department of Microbiology and Infectious Diseases, University of Calgary Health Science Center, for kind consent to use the pMOB3 and pACΩGm vectors for construction of the P. aeruginosa gene-knockout mutants.
REFERENCES
- 1.Brözel, V. S., and T. E. Cloete. 1994. Resistance of Pseudomonas aeruginosa to isothiazolone. J. Appl. Bacteriol. 76:576-582. [DOI] [PubMed] [Google Scholar]
- 2.Davanloo, P., A. H. Rosenberg, J. J. Dunn, and F. W. Studier. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 81:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guérin-Méchin, L., F. Dubois-Brissonnet, B. Heyd, and J. Y. Leveau. 1999. Specific variations of fatty acid composition of Pseudomonas aeruginosa ATCC 15442 induced by quaternary ammonium compounds and relation with resistance to bactericidal activity. J. Appl. Microbiol. 87:735-742. [DOI] [PubMed] [Google Scholar]
- 4.Guérin-Méchin, L., F. Dubois-Brissonnet, B. Heyd, and J. Y. Leveau. 2000. Quaternary ammonium compound stresses induce specific variations in fatty acid composition of Pseudomonas aeruginosa. Int. J. Food Microbiol. 55:157-159. [DOI] [PubMed] [Google Scholar]
- 5.Jones, M. V., T. M. Herd, and H. J. Christie. 1989. Resistance of Pseudomonas aeruginosa to amphoteric and quaternary ammonium biocides. Microbios 58:49-61. [PubMed] [Google Scholar]
- 6.Kourai, H., T. Horie, K. Takeichi, and I. Shibasaki. 1980. The antimicrobial characteristics of quaternary ammonium salts and their alkyl chain length. J. Antibact. Antifung. Agents 8:191-199. [Google Scholar]
- 7.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 8.Li, X., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loughlin, M. F., M. V. Jones, and P. A. Lambert. 2002. Pseudomonas aeruginosa cells adapted to benzalkonium chloride show resistance to other membrane-active agents but not to clinically relevant antibiotics. J. Antimicrob. Chemother. 49:631-639. [DOI] [PubMed] [Google Scholar]
- 10.Maeda, T., S. Goto, Y. Manabe, K. Okazaki, H. Nagamune, and H. Kourai. 1996. Bactericidal action of N-alkylcyanopyridinium bromides against Escherichia coli K12 W3110. Biocontrol Sci. 1:41-49. [Google Scholar]
- 11.Méchin, L., F. Dubois-Brissonnet, B. Heyd, and J. Y. Leveau. 1999. Adaptation of Pseudomonas aeruginosa ATCC 15442 to didecyldimethylammonium bromide induces changes in membrane fatty acid composition and in resistance of cells. J. Appl. Microbiol. 86:859-866. [DOI] [PubMed] [Google Scholar]
- 12.Nagamune, H., C. Ohnishi, A. Katsuura, K. Fushitani, R. A. Whiley, A. Tsuji, and Y. Matsuda. 1996. Intermedilysin, a novel cytotoxin specific for human cells, secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect. Immun. 64:3093-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima, A., M. Hoshikawa, and T. Nakae. 1998. Antibiotic stress induces a large amount of outer membrane protein in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 165:261-265. [DOI] [PubMed] [Google Scholar]
- 14.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-388. [DOI] [PubMed] [Google Scholar]
- 15.Robles, J., and M. Doers. 1994. Promega Notes 45:19. [Google Scholar]
- 16.Russell, A. D. 1998. Bacterial resistance to disinfectants: present knowledge and future problems. J. Hosp. Infect. 43:S57-S68. [DOI] [PubMed] [Google Scholar]
- 17.Russell, A. D. 2000. Do biocides select for antibiotic resistance? J. Pharm. Pharmacol. 52:227-233. [DOI] [PubMed] [Google Scholar]
- 18.Russell, A. D., and G. W. Gould. 1988. Resistance of Enterobacteriaceae to preservatives and disinfectants. J. Appl. Bacteriol. 64:167S-195S. [PubMed]
- 19.Sakagami, Y., H. Yokoyama, H. Nishimura, Y. Ose, and T. Tashima. 1989. Mechanism of resistance to benzalkonium chloride by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 55:2036-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 22.Schweizer, H. P. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-833. [PubMed] [Google Scholar]
- 23.Simon, R., M. O'Connell, M. Labes, and A. Puhler. 1986. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 118:640-659. [DOI] [PubMed] [Google Scholar]
- 24.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, T., T. Murai, I. Fukuda, T. Tobe, M. Yoshikawa, and C. Sasakawa. 1994. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 11:31-41. [DOI] [PubMed] [Google Scholar]
- 26.Tabata, A., T. Maeda, H. Nagamune, and H. Kourai. 2002. Characterization of Pseudomonas aeruginosa resistant to a quaternary ammonium compound. Biocontrol Sci. 7:147-153. [Google Scholar]
- 27.Tattawasart, U., J. Y. Maillard, J. R. Furr, and A. D. Russell. 1999. Development of resistance to chlorhexidine diacetate and cetylpyridinium chloride in Pseudomonas stutzeri and changes in antibiotic susceptibility. J. Hosp. Infect. 42:219-229. [DOI] [PubMed] [Google Scholar]
- 28.Tattawasart, U., J.-Y. Maillard, J. R. Furr, and A. D. Russell. 2000. Outer membrane changes in Pseudomonas stutzeri resistant to chlorhexidine diacetate and cetylpyridinium chloride. Int. J. Antimicrob. Agents 16:233-238. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, L., J. -Y. Maillard, R. J. W. Lambert, and A. D. Russell. 2000. Development of resistance to chlorhexidine diacetate in Pseudomonas aeruginosa and the effect of a ′residual' concentration. J. Hosp. Infect. 46:297-303. [DOI] [PubMed] [Google Scholar]
- 30.Winder, C. L., I. S. I. Al-Adham, S. M. A. Abdel Malek, T. E. J. Buultjens, A. J. Horrocks, and P. J. Collier. 2000. Outer membrane protein shifts in biocide-resistant Pseudomonas aeruginosa PAO1. J. Appl. Microbiol. 89:289-295. [DOI] [PubMed] [Google Scholar]
- 31.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]