Abstract
We describe a program (and a website) to reformat the ClustalX/ClustalW outputs to a format that is widely used in the presentation of sequence alignment data in SNP analysis and molecular systematic studies. This program, CLOURE, CLustal OUtput REformatter, takes the multiple sequence alignment file (nucleic acid or protein) generated from Clustal as input files. The CLOURE-D format presents the Clustal alignment in a format that highlights only the different nucleotides/residues relative to the first query sequence. The program has been written in Visual Basic and will run on a Windows platform. The downloadable program, as well as a web-based server which has also been developed, can be accessed at http://imtech.res.in/~anand/cloure.html.
INTRODUCTION
The ClustalX/ClustalW program is widely used for both protein and nucleic acid multiple sequence alignments and the preparation of phylogenetic trees (1,2). It is freely available, portable and easy to use. The program has undergone many improvements since CLUSTAL was first described in 1988 (3), and is available for different platforms including, most recently, ClustalX. It has an interface to Unix X Windows, the Macintosh MacOS system and MS Windows systems (4,5).
The Clustal program is also widely used in molecular systematics. Ribosomal RNA sequences and their intergenic regions of either bacterial, yeast or other organisms are aligned using Clustal to identify the differences at the genus, species and strain level. These data are often presented as alignments (where the differences are highlighted) or are eventually used to draw phylogenetic trees.
DEVELOPMENT
Despite the extensive use and the advantages of using the Clustal program for nucleic acid alignments in SNP analysis and molecular systematic studies, the program is limited in the options of different formats for presentation. Different software are now available (Jalview, Genedoc) that allow further editing of these sequences (6,7). However, these programs also lack the required format options needed in molecular systematics. In the present manuscript, we have tried to fill this lacunae and have developed a simple, portable (as well as web-based program) that will assist in the presentation of multiple sequence alignment data from Clustal. The program, CLOURE, has been developed for this purpose.
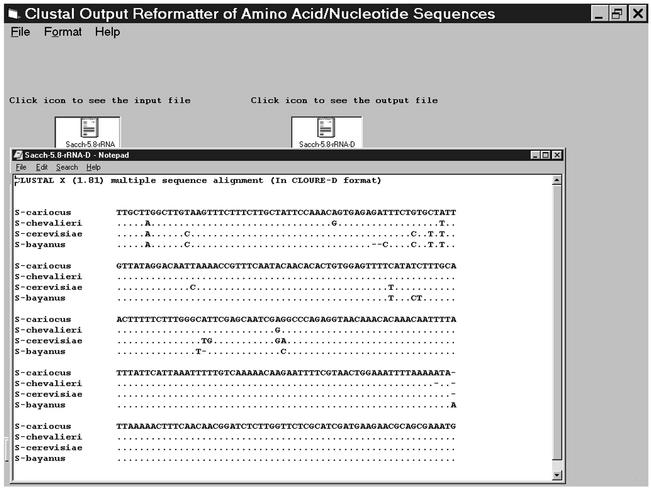
The output of the clustal alignments contains all the loaded sequences that have been aligned according to the parameters of the algorithm defined by the user (Fig. 1). The output format displays the complete sequence of all the sequences selected with a consensus line that includes a ‘*’ for the consensus sequence. In molecular systematics, however, there is a need to highlight the differences, rather than the consensus. The data are most often represented after manual editing of the alignment to highlight only the differences with the parent sequence. The identical residues are represented by a ‘.’ in all the sequences while the difference residues are represented by the appropriate residues. Gaps are represented as ‘-’. As an alternative to manual editing, which can be extremely tedious when one is dealing with a number of sequences, we have developed the CLOURE program: CLustal OUtput REformatter. This program takes the Clustal output as the input (Fig. 1) and after the ‘CLOURE-D’ program is run, it displays the output in the format normally desired for highlighting differences between sequences. A second option, ‘CLOURE-C’ is also available in this program where, once again, the clustal output is taken as an input and in the consensus line of the clustal ouput, the ‘*’ residue is replaced by the single letter code for the amino acid/nucleotide that has been conserved. This is another much favoured presentation for proteins that is also often done manually.
Figure 1.
The CLOURE-D output of a Clustal multiple sequence alignment of the DNA of the 5.8SrRNA region of different Saccharomyces spp. (that acts as the input file for CLOURE). The ouput file (a text file) is clicked to display the contents.
CONCLUDING REMARKS
The CLOURE program has been written in visual basic and has been packaged into a single zip file and can be downloaded from http://imtech.res.in/~anand/cloure.html. Separate versions are available for the different versions of Windows. A web-based program is also available at the site for on-line reformatting.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr G.S. Prasad and Ms Rajeshwari Sutar for helpful suggestions. A.K.B. was supported by a Grant-in-Aid from the Department of Biotechnology, Government of India.
REFERENCES
- 1.Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]
- 2.Jeannmougin F., Thompson,J.D., Gouy,M., Higgins,D.G. and Gibson,T.J. (1998) Multiple sequence alignment with Clustal X. Trends Biochem. Sci., 23, 403–405. [DOI] [PubMed] [Google Scholar]
- 3.Higgins D.G. and Sharp,P.M. (1988) CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene, 73, 237–244. [DOI] [PubMed] [Google Scholar]
- 4.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholas K.B., Nicholas,H.B.,Jr and Deerfield D.W.,II (1997) GeneDoc: analysis and visualization of genetic variation. Embnet News, 4, 1–4. [Google Scholar]
- 7.Clamp M.E., Cuff,J.A. and Barton,G.J. (1998) Jalview: analysis and manipulation of multiple sequence alignments. Embnet News, 5.4. [Google Scholar]