Abstract
3D-Jury is a fully automated protein structure meta prediction system accessible via the Meta Server interface (http://BioInfo.PL/Meta). This is one of the meta predictors, which have made a dramatic, unprecedented impact on the last CASP-5 experiment. The 3D-Jury is comparable with other meta servers but it has the highest combined specificity and sensitivity. The presented method is also very simple and versatile and can be used to create meta predictions even from sets of models produced by humans. An additional and very important and novel feature of the system is the high correlation between the reported confidence score and the accuracy of the model. The number of correctly predicted residues can be estimated directly from the prediction score. The high reliability of the method enables any biologist to submit a target of interest to the Meta Server and screen with relatively high confidence, whether the target can be predicted by fold recognition methods while being unpredictable using standard approaches like PSI-Blast. This can point to interesting relationships which could have been missed in annotations of proteins or genomes and provide very valuable information for novel scientific discoveries.
INTRODUCTION
Protein structure prediction is a mature scientific field with clear application in molecular biology. The structure prediction community evaluates the progress in this field by conducting rigorous, objective, biannual assessment CASP (Critical Assessment of Techniques for Protein Structure Prediction) (1) and CAFASP (Critical Assessment of Fully Automated Structure Prediction) (2) experiments. An important, and probably the main, result of the last round of these experiments (CASP-5 and CAFASP-3) conducted in 2002 is that fully automated structure prediction servers are becoming robust enough to compete with the best human groups. The latest progress is mainly attributed to the development of meta predictors (meta servers), which extract common structural motifs from the set of 3D models generated by various independent prediction providers. The resulting final models have a higher chance of being correct than the models produced by any single method. An important additional advantage of the meta predictors is the improved estimation of the reliability of the predictions. The analysis of the confidence of blind predictions was not conducted successfully so far for human groups, while it is always an important parameter in the assessment of servers.
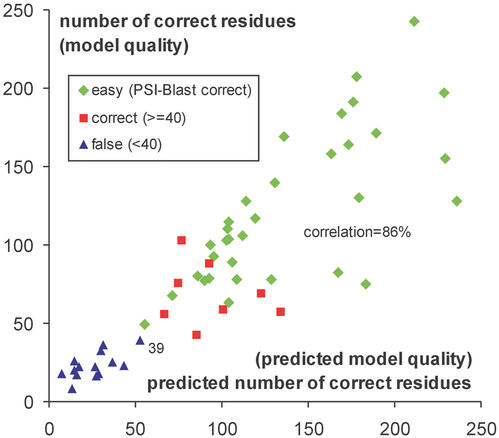
As a result of the progress, users of meta predictors can obtain high quality models but with a negligible fraction of the effort invested usually in predictions by human groups participating in CASP. The results also contain a reliable confidence score, which for one of the best performing meta servers, 3D-Jury, turned out to correlate astonishingly well with the accuracy of the model. Figure 1 shows the correlation obtained for the 3D-Jury meta predictor on CASP-5 targets.
Figure 1.
Correlation between predicted and observed quality of models obtained with 3D-Jury. The x-axis shows the confidence scores reported by 3D-Jury with default settings for all 55 CASP-5 targets with currently known structure. The servers used by the system for consensus building included: ORFeus (11), SamT02 (12), FFAS03 (13), mGenTHREADER (14), INBGU (15), RAPTOR (16), FUGUE-2 (17) and 3D-PSSM (18). The confidence score equals the average number of corresponding residues (residues that can be superimposed on the model) within a selected set of original models provided by other servers. Surprisingly, this number correlates very well with the accuracy of the 3D-Jury model (the number of residues of the model that can be correctly superimposed on the native structure). The y-axis shows the number of C-alpha positions of the model that can be superimposed on the correct structure of the target within 3 Å deviation (correct residues). The correlation between the two values is 86%. Usually, models with 40 or more correct residues are regarded as correct [prediction templates have 90% chance to belong to the same SCOP (19) fold as the targets]. Based on this definition, the points on the plot are divided into 14 false predictions (triangles), eight correct predictions (squares) and 33 correct but easy predictions (rhombs, where PSI-Blast has also generated correct models). 3D-Jury has generated ∼25% more correct predictions than PSI-Blast (PDB-Blast). Only one false prediction (39 correct residues, which is just below the limit for correct predictions) out of 42 has been generated with the default confidence threshold of 50.
DESCRIPTION AND APPLICATION
The 3D-Jury system is a simple and versatile program. The technical details of the algorithm have been published elsewhere (3). The 3D-Jury is comparable with other meta predictors like 3D-ShotGun (4) and the Pcons (5) series, but it has the highest combined specificity and sensitivity (Pcons/Pmodeller seems more specific, while a 3D-ShotGun version is sometimes more sensitive) as assessed in the current LiveBench (6,7) experiment (http://BioInfo.PL/LiveBench). The main advantage over other meta predictors is that the user can decide which of the many prediction providers shall be used for consensus building. The program can utilize predictions provided by human groups as well. Accordingly, this feature was used when analyzing models collected from human experts for the Ten Most Wanted experiment (8).
An important feature of 3D-Jury is its ability to highlight predictions, which are reliable but could not be obtained with confidence using simple application of standard homology detection tools such as PSI-Blast (9) (Fig. 1). This can be used to screen predictions for potentially yet undetected cases of structural or functional similarity. Currently, approximately one out of the 20 targets submitted daily to the Meta Server (10) could be classified this way. Unfortunately, due to limited computational resources, high throughput annotations cannot be conducted yet. The performance of meta predictors could promote the utilization of partially underestimated fold recognition methods, which could lead to an increased access to computation resources by algorithm developers.
It has to be stressed that the development of the meta predictors was only possible as a joint effort of the whole protein structure prediction community. The community designs and constantly improves algorithms, which are shared or offered as servers, conducts objective evaluation experiments and produces user-friendly access to its achievements for the much larger community of biologists.
ACCESS
The 3D-Jury system is accessible via the Structure Prediction Meta Server interface (http://BioInfo.PL/Meta). Due to insufficient computational resources available to some structure prediction servers, the access to the 3D-Jury system is limited to 10 predictions per week from any domain. Special restrictions apply for Polish institutions. This policy is subject to change and can be modified after consultation with the administrators.
REFERENCES
- 1.Moult J., Fidelis,K., Zemla,A. and Hubbard,T. (2001) Critical assessment of methods of protein structure prediction (CASP): round IV. Proteins (Suppl. 5), 2–7. [PubMed] [Google Scholar]
- 2.Fischer D., Elofsson,A., Rychlewski,L., Pazos,F., Valencia,A., Rost,B., Ortiz,A.R. and Dunbrack,R.L.,Jr (2001) CAFASP2: the second critical assessment of fully automated structure prediction methods. Proteins (Suppl. 5), 171–183. [DOI] [PubMed] [Google Scholar]
- 3.Ginalski K., Elofsson,A., Fischer,D. and Rychlewski,L. (2003) 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics, in press. [DOI] [PubMed] [Google Scholar]
- 4.Fischer D. (2003) 3D-SHOTGUN: a novel, cooperative, fold-recognition meta-predictor. Proteins, in press. [DOI] [PubMed] [Google Scholar]
- 5.Lundstrom J., Rychlewski,L., Bujnicki,J. and Elofsson,A. (2001) Pcons: a neural-network-based consensus predictor that improves fold recognition. Protein Sci., 10, 2354–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bujnicki J.M., Elofsson,A., Fischer,D. and Rychlewski,L. (2001) LiveBench-1: continuous benchmarking of protein structure prediction servers. Protein Sci., 10, 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bujnicki J.M., Elofsson,A., Fischer,D. and Rychlewski,L. (2001) LiveBench-2: large-scale automated evaluation of protein structure prediction servers. Proteins (Suppl. 5), 184–191. [DOI] [PubMed] [Google Scholar]
- 8.Fischer D., Baker,D. and Moult,J. (2001) We need both computer models and experiments. Nature, 409, 558. [DOI] [PubMed] [Google Scholar]
- 9.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bujnicki J.M., Elofsson,A., Fischer,D. and Rychlewski,L. (2001) Structure prediction meta server. Bioinformatics, 17, 750–751. [DOI] [PubMed] [Google Scholar]
- 11.Ginalski K., Pas,J., Wyrwicz,L.S., von Grotthus,M., Bujnicki,J.M. and Rychlewski,L. (2003) ORFeus: detection of distant homology using sequence profiles and predicted secondary structure. Nucleic Acids Res., 31, 3804–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karplus K., Karchin,R., Barrett,C., Tu,S., Cline,M., Diekhans,M., Grate,L., Casper,J. and Hughey,R. (2001) What is the value added by human intervention in protein structure prediction? Proteins (Suppl. 5), 86–91. [DOI] [PubMed] [Google Scholar]
- 13.Rychlewski L., Jaroszewski,L., Li,W. and Godzik,A. (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci., 9, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones D.T. (1999) GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol., 287, 797–815. [DOI] [PubMed] [Google Scholar]
- 15.Fischer D. (2000) Hybrid fold recognition: combining sequence derived properties with evolutionary information. Pac. Symp. Biocomput., 5, 116–127. [PubMed] [Google Scholar]
- 16.Xu J., Li,M., Lin,G., Kim,D. and Xu,Y. (2003) Protein threading by linear programming. Pac. Symp. Biocomput., 8, 264–275. [PubMed] [Google Scholar]
- 17.Shi J., Blundell,T.L. and Mizuguchi,K. (2001) FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol., 310, 243–257. [DOI] [PubMed] [Google Scholar]
- 18.Kelley L.A., MacCallum,R.M. and Sternberg,M.J. (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol., 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 19.Lo Conte L., Ailey,B., Hubbard,T.J., Brenner,S.E., Murzin,A.G. and Chothia,C. (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res., 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]