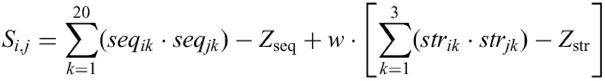Abstract
ORFeus is a fully automated, sensitive protein sequence similarity search server available to the academic community via the Structure Prediction Meta Server (http://BioInfo.PL/Meta/). The goal of the development of ORFeus was to increase the sensitivity of the detection of distantly related protein families. Predicted secondary structure information was added to the information about sequence conservation and variability, a technique known from hybrid threading approaches. The accuracy of the meta profiles created this way is compared with profiles containing only sequence information and with the standard approach of aligning a single sequence with a profile. Additionally, the alignment of meta profiles is more sensitive in detecting remote homology between protein families than if aligning two sequence-only profiles or if aligning a profile with a sequence. The specificity of the alignment score is improved in the lower specificity range compared with the robust sequence-only profiles.
INTRODUCTION
Detection of homology between proteins based on similarity of their sequences can provide a basis for functional predictions for not annotated protein families. However, protein sequences diverge rapidly due to accumulation of amino acid substitutions, hampering the detection of similarity based on pairwise comparisons between many remote homologs. The effectiveness of identification of distant protein relationships has been greatly improved since the introduction of a database search strategy utilizing sequence alignments/profiles as queries instead of simple sequences, as implemented in the broadly used Position-Specific Iterated BLAST (PSI-BLAST) program (1–3). Further significant improvement was made possible owing to the use of sequence profiles for both the query protein and every protein from the database, as implemented in FFAS (4) and in a recently published tool based on information theory (5). ORFeus, the method presented here, incorporates a technique known from fold recognition algorithms. Predicted secondary structure is added to the scoring function, which compares sequence profiles representing potentially homologous protein families. Fold recognition methods (6–9) compare the predicted secondary structure of the query protein with an experimentally determined secondary structure of the protein with known fold. In contrast to this procedure, the predicted secondary structure is used by ORFeus for both the query and the template. This symmetric approach introduced an expected error of over 20% in the description of the secondary structure of the template but has one crucial advantage: protein families with unknown tertiary structure can also be included in the template database. This results in an over 10-fold expansion of the applicability of the algorithm, because most of the known protein sequences lack an experimentally determined structure assignment.
ALGORITHM
The secondary structure prediction is stored in the form of a profile of probabilities. ORFeus can utilize any secondary structure prediction method that produces estimated probabilities for local structure described using three states, that is, the helix, the beta sheet and the loop. Currently the values produced by PSIPRED (10) are used. The sequence profiles are generated as in FFAS (4). The main difference is that all the vectors of probabilities for the occurrence of all amino acids at each position are normalized using the p=1 norm (the sum of all 20 values is equal to 1). The similarity between two positions (elements of the dynamic programming matrix) equals the shifted dot product of the sequence profile vector plus the shifted dot product of the secondary structure probability vector multiplied by the secondary structure weight.
where Sij=score of aligning two positions, seqik=value for the amino acid k in the sequence profile in position i (first protein), seqjk=value for the amino acid k in the sequence profile in position j (second protein), Zseq=zero shift of the sequence profile comparison, strik=value for the secondary structure type k (helix, extended or loop) in the secondary structure profile in position i (first protein), strjk=value for the secondary structure type k (helix, extended or loop) in the secondary structure profile in position j (second protein), Zstr=zero shift of the secondary structure profile comparison and w=secondary structure weight.
The ‘zero shifts’ ensure that the expected score of aligning two positions is below zero. In contrast with FFAS, no normalization of the dynamic programming matrix is conducted. Because of this, the result of the alignment cannot be expressed in normalized scores, but represents only a raw alignment score. FFAS also transforms the alignment score into a Z-score by comparing it to the distribution of alignment scores obtained with a reference databases (both for the query and the template profile), which should additionally increase the accuracy of the final score.
The combined local alignment of two sequence profiles and two secondary structure profiles conducted by ORFeus requires five parameters: gap initiation penalty, gap extension penalty, a weight for the contribution of the secondary structure profiles and two values, which shift the expected dot product of the secondary structure and sequence vectors below zero (expected score of aligning two vectors representing two residues). All five parameters were selected using brute-force optimization on a test set of artificially constructed two-domain families.
The set was based on sequence families extracted from SCOP (11), version 1.55. A set of 472 domains was chosen. The set was divided into two equal groups, so that no fold was represented in both groups (representatives of a fold are either in one or the other group, not in both). In all, 236 proteins in each group were used to create artificial two-domain proteins by concatenating two members (always from different fold classes) into 118 targets. The benchmark of two-domain proteins was used for the development of parameters to reduce the accuracy problem known from earlier FFAS versions, where two-domain proteins had the tendency to be predicted as similar to other unrelated two-domain proteins. The optimization was conducted on one set and the other set was used for the evaluation. A genetic algorithm was used to evolve and improve the parameters. To increase the speed of the optimization the dynamic programming matrices of all 6903 pairs of targets were kept in memory, using a total of 4 Gb RAM on eight dual Pentium® III computers. The new parameters were used only to find the optimal local alignment on a pre-calculated set of dynamic programming matrices.
Two types of scoring functions were used for the optimization of parameters aimed at improving the sensitivity and the specificity of the prediction, respectively. The total sensitivity score for the test set was measured as the sum of prediction scores over all 118 targets. Each prediction score, calculated for each target, is the sum of all correct hits scaled by the number of wrong hits with higher alignment score:
The specificity score was calculated in a similar manner but with all 118·117 alignment scores evaluated simultaneously:
The specificity score enforces higher consistency of alignment scores obtained for different targets. The results of optimizations are presented in Table 1, demonstrating that different parameter sets are selected under different evolutionary pressure. To increase the sensitivity the contribution of the profiles of the predicted secondary structure is increased. At the same time, such a choice results in lowered reliability of the alignment score and it becomes harder to estimate the confidence of the prediction. For comparison, results obtained using only sequence profiles are shown. The scores indicate that the incorporation of the predicted secondary structure improves the accuracy of the prediction method even though the secondary structure profiles are calculated based only on the sequence profiles and do not utilize the experimental data.
Table 1. Optimization results using sensitivity or specificity as scoring function.
| Sequence and 2D profilesa | Sequence profiles onlyb | 2D weightc | Other parametersd | |
|---|---|---|---|---|
| Specificitye | 721 | 705 | 0.06 | 0.08 0.60 0.02 0.63 |
| Sensitivityf | 806 | 749 | 0.30 | 0.08 0.60 0.05 0.70 |
| Limitg | 1038 | 1038 | — |
aScores obtained by the profiles used in ORFeus.
bResults obtained when the predicted secondary structure profiles were not used (2D structure weight equal to zero).
c2D weight: weight of the secondary structure profile when computing the match score between two positions during dynamic programming. The values indicate a 5-fold increase for the importance of the predicted secondary structure when optimized for sensitivity.
dOther parameters: zero shift of the sequence profile score, initiation and extension gap penalty and zero shift of the secondary structure profile score.
eResults obtained using the parameter set optimized for specificity.
fResults obtained using the parameter set optimized for sensitivity.
gThe best theoretically possible results.
The parameters optimized for highest sensitivity were chosen in the final ORFeus implementation, because the improvement of sensitivity over sequence-only profiles was more profound.
IMPLEMENTATION
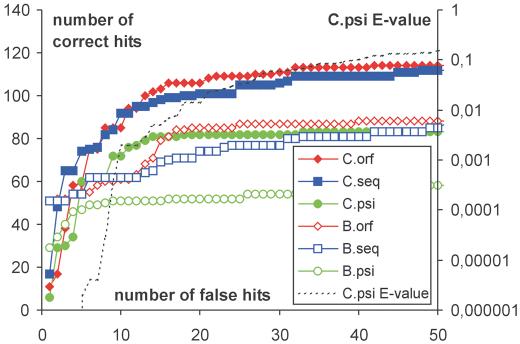
An independent test was conducted on a set containing 1713 family representatives extracted from the current SCOP version 1.57 (representative sequences longer than 600 residues or shorter than 50 residues were removed). Figure 1 shows the number of correctly predicted superfamily relationships as a function of the number of false predictions with higher alignment score. Only one top-scoring prediction for each family is taken into account. This corresponds to the common procedure of specificity evaluation conducted in the LiveBench program (where the evaluated prediction methods use different fold libraries). The performance of ORFeus optimized for sensitivity is compared with a version of the program where only the sequence part of the profile is utilized (optimized with the secondary structure weight equal to zero) and with the PSI-Blast program used also to create the sequence alignments utilized in the profile building procedure. The results show that the more complex meta profiles that utilize predicted secondary structure preferences are more specific than the simple sequence-only profiles in the low specificity range where more than 10% of errors are expected.
Figure 1.
Specificity analysis of profile alignment methods. Three methods: meta-profile alignment ‘orf’ (ORFeus), sequence-only profile alignment ‘seq’ (FFAS-like) and profile with sequence alignment ‘psi’ (PSI-Blast) with two cut-offs for the maximum number of iterations of PSI-Blast (‘B’=3 and ‘C’=6) used to create the sequence profiles are compared (i.e. ‘C.orf’= ORFeus program with six iterations of PSI-Blast used to create the profile). All six procedures were used to predict the superfamily assignment for the 1713 representative sequence of SCOP families. The predictions were sorted by the alignment score. The x-axis shows the number of false predictions (wrong fold assignment) with an alignment score above a cut-off. The left y-axis shows the number of correct predictions (correct superfamily assignment) with an alignment score above a cut-off. The plots show that the sensitivity increases with increasing number of PSI-Blast iterations. With both cut-offs (3 and 6) the profile–profile alignment methods generate in general more correct superfamily assignments than the profile–sequence alignment method (PSI-Blast). The sequence-only alignment methods are more specific in the high specificity range (less than 10 errors), while the meta-profile alignment (ORFeus) generates slightly more correct assignments when more than 10 errors are allowed (specificity <90%). The specificity differences between both profile–profile alignment methods are not very large if compared to PSI-Blast at the same number of iterations. The dotted line (C.psi E-value) shows the E-value (right y-axis) of the false assignment produced by PSI-Blast using six iterations for profile building before scanning the database of 1713 SCOP-family representatives.
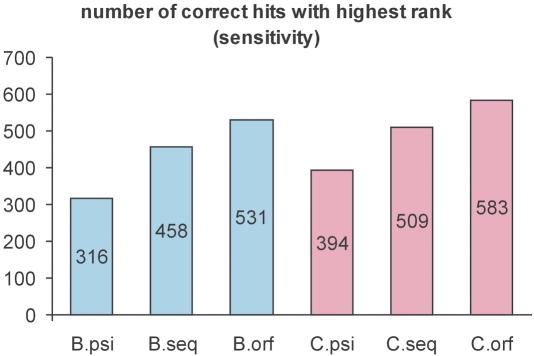
However, the main advantage of meta profiles is their sensitivity. This is demonstrated in Figure 2. The number of correct predictions (with top rank) is plotted for all three prediction procedures. The data confirms again the superiority of aligning sequence profiles over the alignment of a profile with a sequence (PSI-Blast) (12). The alignment of meta profiles conducted by ORFeus is able to boost the sensitivity even further, providing an additional 50% improvement compared with the difference in sensitivity between the other two methods.
Figure 2.
Sensitivity of profile alignment methods. Three methods: meta-profile alignment ‘orf’ (ORFeus), sequence-only profile alignment ‘seq’ (FFAS-like) and profile with sequence alignment ‘psi’ (PSI-Blast) with two cut-offs for the maximum number of iterations of PSI-Blast (‘B’=3 and ‘C’=6) used to create the sequence profiles are compared (i.e. ‘C.orf’= ORFeus program with six iteration of PSI-Blast used to create the profile). All six procedures were used to predict the superfamily assignment for the 1713 representative sequence of SCOP families. The plots show the total number of correct predictions (correct superfamily assignments) obtained using the evaluated methods. The results confirm that increasing the cut-off for the number of PSI-Blast iterations increases the sensitivity of the profiles. At any cut-off (two are shown) the profile–profile alignment methods are more sensitive than the profile–sequence alignment tool. In this analysis the meta profiles also show much higher sensitivity than the sequence-only profile. In the most sensitive setting (six iterations) the sequence-only profile (‘C.seq’) improve the sensitivity of PSI-Blast by ∼30% (∼100 additional correct assignments). An additional 60% increase of this improvement (∼80 correct hits; 20% of correct PSI-Blast hits) can be achieved when adding predicted secondary structure information to the sequence information in the profiles.
The initial comparison with other prediction methods was conducted using the ToolShop service (13). On the ToolShop-2 set ORFeus ranked second in the total sensitivity evaluation of difficult targets and second in total specificity for all targets. Both results were obtained using parameters optimized for sensitivity in November 2001. The only method showing better performance was a consensus predictor Pcons, which combines results obtained from several fold recognition servers (14). A detailed analysis of the performance is available on the ToolShop pages (13).
Owing to the nature of the ToolShop, the result obtained by a server at a later point in time may be over-optimistic. The quick growth of sequence databases provides an artificial advantage for predictions that are conducted later. A more rigorous evaluation was conducted in the LiveBench program (15,16). The completed fourth session confirmed the utility of the presented method (http://BioInfo.PL/LiveBench/4). In the sensitivity evaluation the ORFeus server was only ranked behind novel consensus methods, which utilize several servers or server components to create a jury prediction. The consensus approach is known to result in increased accuracy compared with individual (single-template) prediction methods. From the individual servers only INBGU (8) showed better results than ORFeus on the difficult (HARD) targets, while no individual server was more sensitive than ORFeus on the easy targets. FFAS (4), the sequence-only profile alignment method, was the only individual server that showed higher specificity than ORFeus (a result expected in agreement with prior test results).
The performance of ORFeus was also confirmed in the last CAFASP-3 (17) blind prediction experiment. In the evaluation of autonomous servers ORFeus (the low PSI-Blast iteration version, ORFeus-B) obtained the first rank in the homology modeling sensitivity category and the third rank in the fold recognition category where the second rank was obtained by a server [SHGU (18)] that utilizes consensus building and fragment splicing technology.
CONCLUSIONS
The addition of predicted secondary structure to conventional sequence profiles is able to boost the sensitivity of profile–profile comparison methods substantially. This addition is, however, accompanied by a serious distortion of the alignment score distribution. The increase of sensitivity should result in an increase of specificity in our benchmarks since more correct predictions are expected in total. This has not happened and the specificity of sequence-only profiles remains on a similar level as that of the meta profiles. In particular, in high specificity ranges the conventional sequence-only profiles remain more robust.
The currently best way to boost the specificity of predictions is the application of consensus methods. ORFeus will become a valuable component of such methods providing a high number of correct family assignments despite limited specificity of the alignment score. ORFeus has already been incorporated in newer Pcons/Pmodeller versions (14).
ACCESS
ORFeus is available to the academic community via the convenient Structure Prediction Meta Server (http://BioInfo.PL/Meta) or through the experimental higher throughput GRDB system pages (http://grdb.bioinfo.pl). A commercial standalone version of the program is available upon request. ORFeus is also coupled to the continuous online server evaluation program, LiveBench (http://BioInfo.PL/LiveBench/).
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by the 5th Framework Programme grant QLK3-CT-2000-00170.
REFERENCES
- 1.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L. and Koonin,E.V. (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 3.Schaffer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf,Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rychlewski L., Jaroszewski,L., Li,W. and Godzik,A. (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci., 9, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yona G. and Levitt,M. (2002) Within the twilight zone: a sensitive profile–profile comparison tool based on information theory. J. Mol. Biol., 315, 1257–1275. [DOI] [PubMed] [Google Scholar]
- 6.Jones D.T. (1999) GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J. Mol. Biol., 287, 797–815. [DOI] [PubMed] [Google Scholar]
- 7.Kelley L.A., MacCallum,R.M. and Sternberg,M.J. (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol., 299, 499–520. [DOI] [PubMed] [Google Scholar]
- 8.Fischer D. (2000) Hybrid fold recognition: combining sequence derived properties with evolutionary information. Pac. Symp. Biocomput., 5, 116–127. [PubMed] [Google Scholar]
- 9.Shi J., Blundell,T.L. and Mizuguchi,K. (2001) FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol., 310, 243–257. [DOI] [PubMed] [Google Scholar]
- 10.Jones D.T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol., 292, 195–202. [DOI] [PubMed] [Google Scholar]
- 11.Lo Conte L., Ailey,B., Hubbard,T.J., Brenner,S.E., Murzin,A.G. and Chothia,C. (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res., 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panchenko A.R. (2003) Finding weak similarities between proteins by sequence profile comparison. Nucleic Acids Res., 31, 683–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rychlewski L. (2001) ToolShop: prerelease inspections for protein structure prediction servers. Bioinformatics, 17, 1240–1241. [DOI] [PubMed] [Google Scholar]
- 14.Lundstrom J., Rychlewski,L., Bujnicki,J. and Elofsson,A. (2001) Pcons: a neural-network-based consensus predictor that improves fold recognition. Protein Sci., 10, 2354–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bujnicki J.M., Elofsson,A., Fischer,D. and Rychlewski,L. (2001) LiveBench-1: continuous benchmarking of protein structure prediction servers. Protein Sci., 10, 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bujnicki J.M., Elofsson,A., Fischer,D. and Rychlewski,L. (2001) LiveBench-2: large-scale automated evaluation of protein structure prediction servers. Proteins (Suppl. 5), 184–191. [DOI] [PubMed] [Google Scholar]
- 17.Fischer D., Elofsson,A., Rychlewski,L., Pazos,F., Valencia,A., Rost,B., Ortiz,A.R. and Dunbrack,R.L.,Jr (2001) CAFASP2: the second critical assessment of fully automated structure prediction methods. Proteins (Suppl. 5), 171–183. [DOI] [PubMed] [Google Scholar]
- 18.Fischer D. (2003) 3D-SHOTGUN: A novel, cooperative, fold-recognition meta-predictor. Proteins, in press. [DOI] [PubMed] [Google Scholar]