Abstract
We present a coherent series of servers that can perform a large number of structure analyses on nuclear hormone receptors. These servers are part of the NucleaRDB project, which provides a powerful information system for nuclear hormone receptors. The computations performed by the servers include homology modelling, structure validation, calculating contacts, accessibility values, hydrogen bonding patterns, predicting mutations and a host of two- and three-dimensional visualisations. The Nuclear Receptor Structure Analysis Servers (NRSAS) are freely accessible at http://www.cmbi.kun.nl/NR/servers/html/ and in-house copies can be obtained upon request.
INTRODUCTION
Complicated data analysis tools are rapidly becoming an integral aspect of work in most bioscience laboratories. Unfortunately, many tools are expensive, difficult to install or use, require special hardware or are otherwise not easily accessible to scientists. The World Wide Web (WWW) is a good solution for this problem. Specialists can maintain WWW-based servers in laboratories that have adequate hardware, software and expertise available, while the scientists only have to concentrate on the scientific aspects of servers.
Nuclear hormone receptors (NRs) are becoming a very important topic for the pharmaceutical industry, which is spurring a flurry of activities. The NucleaRDB (1) was set up to gather, combine and disseminate information on NRs. This information system integrates sequence data, mutation data, ligand binding data and three-dimensional structures to form a one-stop-shopping-centre for NR-related information.
WHAT IF (2) is a versatile program for the analysis, manipulation, prediction and visualisation of macromolecular structures as stored in the PDB (3) databank. This program is very widely used, in particular by research groups interested in different aspects of protein structure analysis. Unfortunately, the large number of options and commands that WHAT IF offers makes it difficult to use and limits its applicability. This is a problem for laboratories that only occasionally need to analyse protein structures. We have solved this problem for the users of the NucleaRDB by producing a series of WWW-based servers that can run many WHAT IF options on nuclear receptor structures.
Table 1 lists some of the options that can be performed by the Nuclear Receptor Structure Analysis Servers (NRSAS). Figure 1 shows the WWW page for a typical server. The NRSAS are freely available at http://www.cmbi.kun.nl/NR/servers/html/.
Table 1. Server classes and descriptions.
| Server class | Server description |
|---|---|
| Administration | List residue |
| List sequence | |
| List coordinates | |
| Renumber a file | |
| Structure validation | Check packing quality |
| Check Ramachandran plot | |
| Check crystal packing clashes | |
| Check bond angles (or distances, planarities, etc.) | |
| Protein analysis | Accessibility, secondary structure and crystal contacts |
| Protein ligand contacts | |
| 2D graphics | B-factor plot |
| Ramachandran plot | |
| 3D graphics | Molecule coloured by B-factor |
| Transparent arrow-and-cylinder drawing | |
| A simple alpha carbon trace | |
| Cysteine related | List cysteine bridges |
| List torsion angles of cysteine bridges | |
| Water related | List waters near residues |
| List residues near waters | |
| Move waters to the correct symmetry position | |
| Add crystallographically related waters missing in PDB file |
About 100 servers are divided into 20 classes, approximately.
The left column lists some of these classes, while the right column lists a few servers of each of those classes.
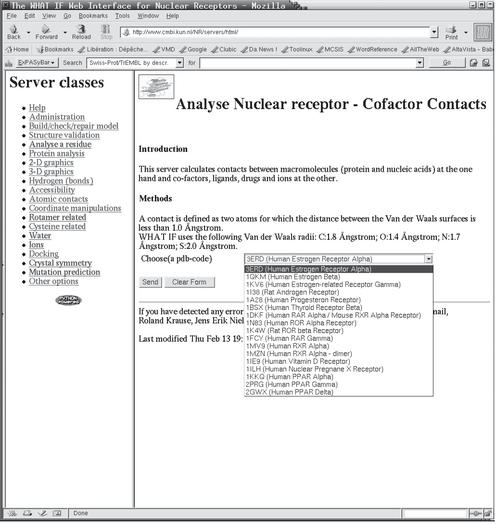
Figure 1.
NRSAS input screenshot. The right frame is the input form of the server that calculates protein–ligand interactions. The pull-down menu with 17 representative nuclear receptor related PDB files is activated. The left frame lists all 19 classes of options. (See Fig. 2 for the result of activating this server.)
STRUCTURE AND USE OF SERVERS
All the servers, input forms, result pages, and so on, are produced by a python script. This script reads a meta-file in which the server manager describes in English the tasks the server should execute. This meta-file also contains the help texts and explanations for all the input and output forms and tables. A script for the program WHAT IF forms the most complicated part of the meta-file. This script contains all the WHAT IF commands a user would have typed in an interactive session. File names, residue ranges, and so on, have to be indicated by the user. The server builder therefore knows a series of commands that it can extract from the WHAT IF script, and converts it into input boxes in the WWW input form. Upon execution, the values typed by the user in those input boxes will be inserted in the WHAT IF script. This generates a valid WHAT IF script that performs the desired option with no need for the user to understand the script language and the interactive options of WHAT IF.
It is therefore possible to make one's own WHAT IF based servers. Only a WHAT IF license and the python scripts to build and maintain the servers are needed. A basic understanding of the WHAT IF software to write WHAT IF scripts is also required. Python (1.5 or higher), WHAT IF and an HTTP server (e.g. Apache) have to be installed prior to setting up and running the servers. Installation notes and other information are available via the WWW pages for the servers.
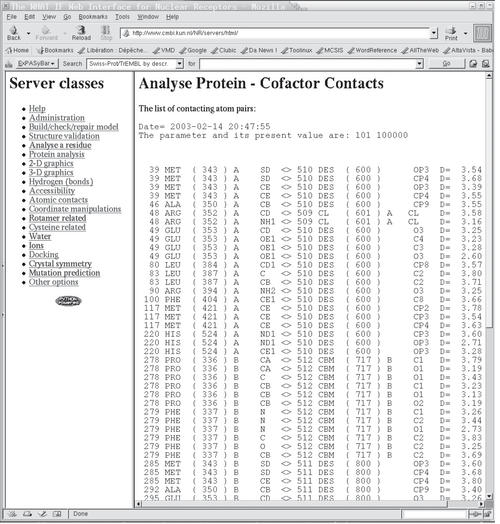
Figure 1 shows a screenshot of a typical server page. This server lists the contacts between the protein and ligands. Figure 2 shows a screenshot of the output obtained a few seconds after submitting the job prepared in Figure 1.
Figure 2.
NRSAS output screenshot. The left frame lists all 19 classes of options. The right frame shows the results of the server run, as illustrated in Figure 1. From left to right the residue number, residue type, PDB identifier of residue, chain identifier and name of atom making the contact are indicated. The left five columns are for the atom of the macromolecule; the next five columns are for the atom of the ligand. The right column gives the inter-atomic distance in Ångstrom.
It takes about 30 seconds to build 100 servers. We therefore do not provide options to add or delete servers. Servers are updated by editing the server meta-file and running the server generation script again. Figure 3 shows the part of the meta-file needed by the server shown in Figures 1 and 2.
Figure 3.
Section of the meta-file used to build the server illustrated in Figures 1 and 2. Most data can easily be correlated to the content of the forms in Figures 1 and 2. The input record sets up the chooser for nuclear receptor PDB files. The six lines that start with ‘getmol’ correspond to a WHAT IF script.
We believe that this concept of servers for molecular class specific protein structure analysis will be useful for scientists who like to have occasional access to such software but lack the time, money, software, hardware or human infrastructure to maintain such facilities in their laboratory.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank their colleagues at the CMBI, Organon and Unilever for stimulating discussions. Rob Hooft and Jens Nielsen provided valuable technical support. This project was supported by LionBiosciences Ag in Heidelberg, Germany.
REFERENCES
- 1.Horn F., Vriend,G. and Cohen,F.E. (2001) Collecting and harvesting biological data: the GPCRDB and NucleaRDB information systems. Nucleic Acids Res., 29, 346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vriend G. (1990) WHAT IF: a molecular modelling and drug design program. J. Mol. Graph., 8, 52–56. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein F.C., Koetzle,T.F., Williams,G.J.B., Meyer,E.F.,Jr, Brice,M.D., Rogers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]