Abstract
The detection of local structural patterns in proteins (e.g. active sites) can provide insights into protein function in the absence of sequence or fold similarity. Methods to detect such similarities are key during structural annotation, for example with results from Structural Genomics initiatives. PINTS (Patterns in Non-homologous Tertiary Structures, http://pints.embl.de) performs database searches for such patterns and most importantly provides a measure of statistical significance for any similarity uncovered. To aid functional annotation of proteins, we allow comparisons of pre-defined patterns against databases of complete structures and of entire structures to databases of particular residues likely to be functionally important.
INTRODUCTION
Recent improvements in structural biology have greatly increased the number of protein three-dimensional (3D) structures (1). These have culminated in Structural Genomics projects (2), which aim to solve the structures for all proteins as a means to understanding function. Methods to annotate function through structure are thus now of growing importance (3,4).
Proteins of known structure, but of unknown function are typically compared to databases of other structures to discover functional relationships. One class of methods such as DALI (5,6), VAST (7), SSAP (8) or STAMP (9) compare structures to a database using alignments, and thus find proteins with a similar fold (common spatial arrangements of secondary structure elements in the same order along the sequence). Such similarities can identify ancient evolutionary relationships that are not always apparent when only sequences are known, but that are often associated with a similarity in function. Indeed, the location of active sites or binding surfaces or substrate type is often conserved and their function can be easily tested by further experiments (10–20).
However, search methods based on structural alignment do not always provide functional clues. This is clear if a protein adopts a new fold (i.e. does not resemble any known structure), but problems can also arise when proteins adopt very common folds that perform many different functions, such as β/α-(TIM)-barrel, ferredoxin or immunoglobulin-like structures (21). Here functional inferences are difficult to make, as structural alignments can show an equal degree of similarity between functionally similar and dissimilar proteins.
An alternative strategy is to obtain functional clues by detecting local structural patterns associated with a particular function, which can be common to proteins with different folds. Residues within these patterns are not necessarily adjacent in the protein sequence and can occur in any order. A classic example of this phenomenon is the trypsin-like catalytic triad, which nature has reinvented more than ten times (22), although several other instances have been reported (23–25). These functionally important similarities cannot be detected by sequence comparison or structural alignments and require methods that are independent of sequence or fold similarity (23,26–31).
The PINTS (Patterns in Non-homologous Tertiary Structures, http://pints.embl.de) server allows such similarities to be uncovered. Unlike previous methods, it also provides a measure of statistical significance similar to that used by BLAST (32–34). This lets the user easily assess whether a match is likely to have functional implications or is a background match found by chance. PINTS also aids functional annotation of new structures by providing databases with functionally relevant patterns, thus avoiding the need to consider many non-functional (e.g. structural) matches that might arise when comparing entire structures.
ABOUT PINTS
At the heart of the server is a protein structure comparison method that uses a depth-first search method (similar to that described in 23). It finds all possible patterns of residues (or atoms or points defined by other criteria) common to two sets of coordinates, which are both close in space and geometrically similar, as measured by comparison of inter-residue distances.
For each pattern found we calculate the root-mean-square deviation (RMSD) between the matched atoms. RMSD accurately scores the difference between two sets of coordinates and is often used in structure comparison [as for example in SPASM (29,35), which allows the comparison of patterns to databases of protein structures]. However, the RSMD that implies a meaningful similarity is highly dependent on the number and type of atoms being compared. To avoid ambiguities, or the choice of an arbitrary RMSD cut-off for any particular pattern, we provide a measure of statistical significance based on a rigorous model for the behavior of RMSD (36). PINTS gives E-values similar to those used in sequence searches that assess the probability that the obtained matches occurred just by chance without further functional implications (32,33). Measures of statistical significance also mean that matches with different numbers of amino acid residues and atoms can be easily compared across different searches, and permit a single cut-off to be applied. RMSD alone can place insignificant matches with fewer residues/atoms above those that are larger and significant, in spite of a higher RMSD. It is this feature that sets PINTS apart from previously used methods (23,26–31) or servers (35) that perform such searches.
THE PINTS SERVER
The PINTS server provides an easy-to-use interface and offers several databases of complete protein structures or patterns (Table 1). Search results are kept for eight days and can be retrieved by an identifier, Email or IP address as preferred. The server currently allows for three types of searches, as illustrated in Figure 1.
Table 1. Databases of complete protein structures or patterns.
| Database | Description |
|---|---|
| Proteins | |
| SCOP (39) representatives | A representative (the first) taken from each: |
| Folds | Fold |
| SFams | Superfamily |
| Fams | Family |
| P.Species | Protein or Species |
| PDBselect 25 | Chains with <25% sequence identity (46) |
| PDBselect 90 | <90% |
| Patterns | Residues that are: |
| Ligand-binding sites | within 3.0 Å of a HETATM in the PDB |
| SITE annotations | extracted from all annotated SITEs in the PDB |
| Surface residues | more than 25% exposed [DSSP (37)] |
| Conserved residues | conserved in 80% of close homologues (available soon) |
Figure 1.
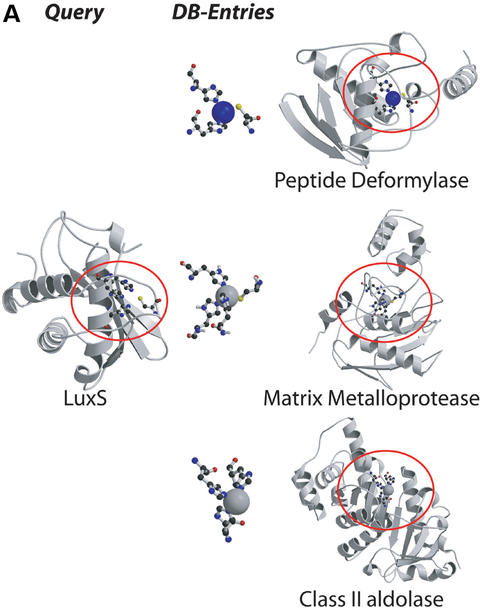
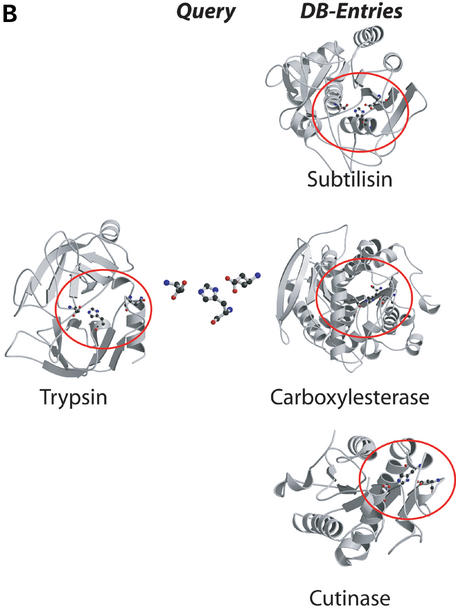
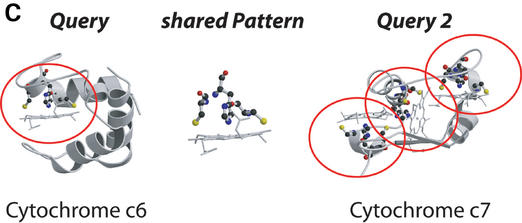
Examples for the three types of searches offered by the PINTS server. (A) Protein versus pattern search: we compared the entire structure of LuxS (PDB entry 1j98) to the database of functional residues defined by the authors (SITE, Table 1). LuxS is shown on the left, and representatives from the top scoring folds on the right [ribbons for α-helices, arrows for β-strands; drawn by Molscript (44) and Raster3D (45)]. Residues found to be similar between LuxS and database entries are circled and shown in ball-and-stick. The representatives (E-values, PDB codes and associated sites) are peptide deformylase (2×10−6, 1bs6, NIC=active site near nickel), the zincin-like matrix-metalloprotease stromelysin-1 (8.3×10−5, 1slm, ZN1=ligands of the catalytic ion) and the class II aldolase rhamnulose-1-phosphate aldolase (0.2, 1gt7, ZNA=Zn binding site). Matches to sites within proteins of the LuxS/MPP-like metallohydrolase (i.e. proteins similar to LuxS), cysteine rich, dioxygenase and classical C2H2,C2HC zinc finger folds are not shown for clarity. (B) Pattern versus protein search: we compared the catalytic triad from trypsin (1mct, left) to one representative of each SCOP family (Table 1). Top scoring matches shown on the left (equivalent patterns circled) are to the subtilisin-like fold (E=0.4, 1cse, subtilisin), the α/β-hydrolases (E=0.8, 1jkm, carboxylesterase) and the flaxodoxin-like cutinases (E=0.8, 1cex, cutinase). (C) Pairwise comparison of entire proteins: we compared cytochrome c6 (1c75) and the multiheme cytochrome c7 (1hh5). Three similarities were detected, all involving the CxxCH heme attachment site, of which one is present in cytochrome c6 (residues 32,35,36) and three in c7 (1: 26,29,30, E=10−6; 2: 49,52,53, E=0.18; 3: 62,65,66, E=3.6×10−4). The match to site 2 also identified an additional serine (residue 44 in c6 and 55 in c7) common to both proteins.
Protein versus pattern database
For a new protein structure, hints about function or the location of a functional site can come from searches against databases of patterns likely to be of functional importance (Table 1). For the ligand binding sites database we collected residues that have at least one atom within 3.0 Å of a HETATM entry (excluding waters). The surface residues database contains all residues with a relative accessibility >25% as measured by DSSP (37) and the SITE annotations database gives those defined by structural biologists as forming a functional site (SITE entries in PDB files).
Pattern versus protein database
The recurrence of a known functional or an interesting new pattern in other structures can suggest common properties. We therefore allow patterns of up to 10 residues to be compared to protein databases (i.e. containing complete structures) at different levels of redundancy (Table 1) or the pattern databases above. The user can either upload patterns in PDB format or select them from larger structures by an easy syntax.
Pairwise comparison
For two proteins that share a biochemical or cellular function (e.g. catalytic activity, specific binding characteristics, etc.) a pairwise comparison, considering the entire structures, can suggest the molecular basis for the common feature. PINTS thus allows a pairwise comparison of two structures that the user can either upload or select from the PDB (1).
We distinguish between the three types of comparisons because a single, all-encompassing, all-against-all search, would greatly increase the search space. This not only affects the CPU time, but has a critical effect on the statistics: searching more amino acids increases the number of random matches, and can have the effect of burying true matches in noise. For example, a protein versus pattern search comparing trypsin to a database of functional patterns, or a pattern versus protein database search comparing only the catalytic triad (1mct: His-57, Asp-102, Ser-195) to a database of whole structures identifies true functional similarities to be significant. However, a pairwise comparison between trypsin and subtilisin detects the similarity, but does not find it to be significant owing to the large number of background matches introduced by the comparison of two whole proteins (of 223 and 275 amino acids, respectively). This is not a limitation of the method, but a fact of life when searching for similarities within large databases (38).
We currently restrict the search parameters to standard settings that we know would be applicable to a wide variety of different submissions (maximum pattern diameter 15 Å, distance tolerance during the depth-first search 3 Å, exclusion of hydrophobic residues from the search, minimum and maximum number of residues per pattern 3 and 10, respectively).
For all searches, matches up to a user-defined E-value maximum and that contain at least three residues are reported. We allow for partial matches to be detected, which is particularly important if an active site is not fully understood or when the similarity may not cover the whole of a pre-defined site. Automatically or manually annotated patterns (such as the ligand binding sites or the SITE annotations database entries in PINTS) often contain additional residues that are not absolutely required for function.
Matches are ranked by their statistical significance and the equivalent residues and associated RMSDs are provided, as are cross-references to useful on-line resources such as PDB (1), SCOP (39), NCBI-Entrez (http://www.ncbi.nlm.nih.gov/Entrez/) and PDBsum (40–42). For visual inspection with RasMol (43), we provide superimposed coordinates for both the matched patterns alone (i.e. the equivalent residues) and within the whole protein context.
BENCHMARKING AND INTERPRETATION OF RESULTS
A detailed benchmark of PINTS is difficult to achieve as there is currently no large reliable resource of similar protein active sites. However, we have tested the method on a large set of known similarities and found that virtually all could be detected with statistical significance (36).
We compared a set of protein structures recently determined by structural genomics projects (3) to a database of residues from proteins of known structure in contact with bound ligands (Table 1). Inspection of the matches revealed that highly similar binding sites had E<10−5, those with similar chemical groups had values in the range of 10−4–10−2 and negatives had E>0.1. Although these values vary with the size of the database used, they can guide interpretation of the results during structural annotation.
FUTURE DEVELOPMENTS
We are constantly updating the PINTS server and aim to serve the structural biology community in the best possible way. Currently, we are adding a pattern database of residues that are conserved in homologous sequences suggesting an important structural or functional role. In addition, we are implementing a BLAST-based filter (34) that detects proteins with high sequence identity and removes them from the search. This filter will enable us to allow searches with entire structures against protein databases. We will also soon permit searches with patterns that contain non-protein atoms such as functionally important water molecules. As our experience grows with the users' needs, we will also allow more freedom in defining parameters such as conservative amino acid substitutions, or those related to pattern dimensions restricted during the depth-first search algorithm.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Caroline Hadley and Hugo Ceulemans (EMBL) for a critical reading of the manuscript and helpful comments on the server.
Note added in proof
The BLAST-based filter is now in place, and we currently allow searches with proteins of up to 100 amino acids against databases of whole structures.
REFERENCES
- 1.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The protein data bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burley S.K. (2000) An overview of structural genomics. Nature Struct. Biol., 7, 932–934. [DOI] [PubMed] [Google Scholar]
- 3.Teichmann S.A., Murzin,A.G. and Chothia,C. (2001) Determination of protein function, evolution and interactions by structural genomics. Curr. Opin. Struct. Biol., 11, 354–363. [DOI] [PubMed] [Google Scholar]
- 4.Thornton J.M., Todd,A.E., Milburn,D., Borkakoti,N. and Orengo,C.A. (2000) From structure to function: approaches and limitations. Nature Struct. Biol., 7, 991–994. [DOI] [PubMed] [Google Scholar]
- 5.Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 6.Holm L. and Sander,C. (1995) Dali: a network tool for protein structure comparison. Trends Biochem. Sci., 20, 478–480. [DOI] [PubMed] [Google Scholar]
- 7.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 8.Orengo C.A. and Taylor,W.R. (1996) SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol., 266, 617–635. [DOI] [PubMed] [Google Scholar]
- 9.Russell R.B. and Barton,G.J. (1992) Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins, 14, 309–323. [DOI] [PubMed] [Google Scholar]
- 10.Chiu H.J., Johnson,E., Schroder,I. and Rees,D.C. (2001) Crystal structures of a novel ferric reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus and its complex with NADP+. Structure (Camb.), 9, 311–319. [DOI] [PubMed] [Google Scholar]
- 11.Colovos C., Cascio,D. and Yeates,T.O. (1998) The 1.8 Å crystal structure of the ycaC gene product from Escherichia coli reveals an octameric hydrolase of unknown specificity. Structure, 6, 1329–1337. [DOI] [PubMed] [Google Scholar]
- 12.Lima C.D., Klein,M.G. and Hendrickson,W.A. (1997) Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science, 278, 286–290. [DOI] [PubMed] [Google Scholar]
- 13.Minasov G., Teplova,M., Stewart,G.C., Koonin,E.V., Anderson,W.F. and Egli,M. (2000) Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP. Proc. Natl Acad. Sci. USA, 97, 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volz K. (1999) A test case for structure-based functional assignment: the 1.2 Å crystal structure of the yjgF gene product from Escherichia coli. Protein Sci., 8, 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F., Gustafson,K.R., Boyd,M.R. and Wlodawer,A. (1998) Crystal structure of Escherichia coli HdeA. Nature Struct. Biol., 5, 763–764. [DOI] [PubMed] [Google Scholar]
- 16.Cort J.R., Yee,A., Edwards,A.M., Arrowsmith,C.H. and Kennedy,M.A. (2000) Structure-based functional classification of hypothetical protein MTH538 from Methanobacterium thermoautotrophicum. J. Mol. Biol., 302, 189–203. [DOI] [PubMed] [Google Scholar]
- 17.Sinha S., Rappu,P., Lange,S.C., Mantsala,P., Zalkin,H. and Smith,J.L. (1999) Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family. Proc. Natl Acad. Sci. USA, 96, 13074–13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du X., Choi,I.G., Kim,R., Wang,W., Jancarik,J., Yokota,H. and Kim,S.H. (2000) Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-A resolution. Proc. Natl Acad. Sci. USA, 97, 14079–14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yee A., Chang,X., Pineda-Lucena,A., Wu,B., Semesi,A., Le,B., Ramelot,T., Lee,G.M., Bhattacharyya,S., Gutierrez,P., et al. (2002) An NMR approach to structural proteomics. Proc. Natl Acad. Sci. USA, 99, 1825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang K.Y., Chung,J.H., Kim,S.H., Han,Y.S. and Cho,Y. (1999) Structure-based identification of a novel NTPase from Methanococcus jannaschii. Nature Struct. Biol., 6, 691–696. [DOI] [PubMed] [Google Scholar]
- 21.Orengo C.A., Jones,D.T. and Thornton,J.M. (1994) Protein superfamilies and domain superfolds. Nature, 372, 631–634. [DOI] [PubMed] [Google Scholar]
- 22.Dodson G. and Wlodawer,A. (1998) Catalytic triads and their relatives. Trends Biochem. Sci., 23, 347–352. [DOI] [PubMed] [Google Scholar]
- 23.Russell R.B. (1998) Detection of protein three-dimensional side-chain patterns: new examples of convergent evolution. J. Mol. Biol., 279, 1211–1227. [DOI] [PubMed] [Google Scholar]
- 24.Denessiouk K.A., Lehtonen,J.V., Korpela,T. and Johnson,M.S. (1998) Two ‘unrelated’ families of ATP-dependent enzymes share extensive structural similarities about their cofactor binding sites. Protein Sci., 7, 1136–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endicott J.A. and Nurse,P. (1995) The cell cycle and suc1: from structure to function? Structure, 3, 321–325. [DOI] [PubMed] [Google Scholar]
- 26.Artymiuk P.J., Poirrette,A.R., Grindley,H.M., Rice,D.W. and Willett,P. (1994) A graph-theoretic approach to the identification of three-dimensional patterns of amino acid side-chains in protein structures. J. Mol. Biol., 243, 327–344. [DOI] [PubMed] [Google Scholar]
- 27.Fetrow J.S. and Skolnick,J. (1998) Method for prediction of protein function from sequence using the sequence-to-structure-to-function paradigm with application to glutaredoxins/thioredoxins and T1 ribonucleases. J. Mol. Biol., 281, 949–968. [DOI] [PubMed] [Google Scholar]
- 28.Fischer D., Wolfson,H., Lin,S.L. and Nussinov,R. (1994) Three-dimensional, sequence order-independent structural comparison of a serine protease against the crystallographic database reveals active site similarities: potential implications to evolution and to protein folding. Protein Sci., 3, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleywegt G.J. (1999) Recognition of spatial motifs in protein structures. J. Mol. Biol., 285, 1887–1897. [DOI] [PubMed] [Google Scholar]
- 30.Wallace A.C., Borkakoti,N. and Thornton,J.M. (1997) TESS: a geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci., 6, 2308–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1996) Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Sci., 5, 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul S.F. and Gish,W. (1996) Local alignment statistics. Methods Enzymol., 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 33.Karlin S. and Altschul,S.F. (1990) Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl Acad. Sci. USA, 87, 2264–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaffer A.A., Aravind,L., Madden,T.L., Shavirin,S., Spouge,J.L., Wolf,Y.I., Koonin,E.V. and Altschul,S.F. (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res., 29, 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madsen D. and Kleywegt,G.J. (2002) Interactive motif and fold recognition in protein structures. J. Appl. Cryst., 35, 137–139. [Google Scholar]
- 36.Stark A., Sunyaev,S. and Russell,R.B. (2003) A model for statistical significance of local similarities in structure. J. Mol. Biol., 326, 1307–1316. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 38.Jones D.T. and Swindells,M.B. (2002) Getting the most from PSI-BLAST. Trends Biochem. Sci., 27, 161–164. [DOI] [PubMed] [Google Scholar]
- 39.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski R.A. (2001) PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res., 29, 221–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luscombe N.M., Laskowski,R.A., Westhead,D.R., Milburn,D., Jones,S., Karmirantzou,M. and Thornton,J.M. (1998) New tools and resources for analysing protein structures and their interactions. Acta Crystallogr. D Biol. Crystallogr., 54, 1132–1138. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski R.A., Hutchinson,E.G., Michie,A.D., Wallace,A.C., Jones,M.L. and Thornton,J.M. (1997) PDBsum: a Web-based database of summaries and analyses of all PDB structures. Trends Biochem. Sci., 22, 488–490. [DOI] [PubMed] [Google Scholar]
- 43.Sayle R.A. and Milner-White,E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 374–376. [DOI] [PubMed] [Google Scholar]
- 44.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst., 24, 946–950. [Google Scholar]
- 45.Merritt E.A. and Murphy,M.E.P. (1994) Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D Biol. Crystallogr., 50, 869–873. [DOI] [PubMed] [Google Scholar]
- 46.Hobohm U. and Sander,C. (1994) Enlarged representative set of protein structures. Protein Sci., 3, 522–524. [DOI] [PMC free article] [PubMed] [Google Scholar]