Abstract
PredictProtein (PP, http://cubic.bioc.columbia.edu/pp/) is an internet service for sequence analysis and the prediction of aspects of protein structure and function. Users submit protein sequence or alignments; the server returns a multiple sequence alignment, PROSITE sequence motifs, low-complexity regions (SEG), ProDom domain assignments, nuclear localisation signals, regions lacking regular structure and predictions of secondary structure, solvent accessibility, globular regions, transmembrane helices, coiled-coil regions, structural switch regions and disulfide-bonds. Upon request, fold recognition by prediction-based threading is available. For all services, users can submit their query either by electronic mail or interactively from World Wide Web.
OVERVIEW
Ten years of sustained service for protein structure prediction
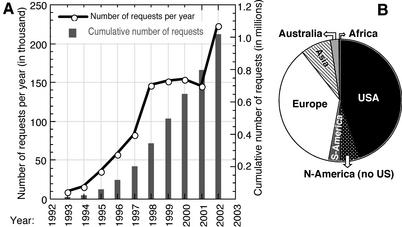
PredictProtein (PP) is an automatic service that searches up-to-date public sequence databases, creates alignments and predicts aspects of structure and function (Fig. 1). Users send a protein sequence and receive a single file with results from database comparisons and prediction methods. PP went online in 1992 at the European Molecular Biology Laboratory (EMBL, Heidelberg) (1); it was the first internet server for protein structure prediction, and belonged to a group of five pioneering internet sites for molecular biology (2). Originally, PP handled all requests through email. Since summer 1993, users can also query the server through a web interface and opt for an HTML output format. When the server moved from EMBL to Columbia University (1999), this output format was extended significantly and we added the option of retrieving the results through http download to account for the continuously growing sizes of alignments. With the explosion of the web in the mid 90s, many other servers have implemented particular aspects covered by PP. However, PP remains the most widely used public server for structure prediction: over one million requests from users in 95 countries have been handled over the first decade of PP (Fig. 2A). Ten or more different proteins were submitted by 11 110 users. About 45% of the requests originated from the USA, 78% from North America and Europe (Fig. 2B). PP web pages are mirrored in 17 countries on four continents. Despite this load-spread, the PP pages at New York alone were listed as one of the most frequently accessed internet sites in bioinformatics by ‘Links2go’ with over 17 000 daily hits (>3 million since January 2002). Our goal has always been to develop a system optimised to meet the demands of experimentalists not experienced in bioinformatics. This implied that we focused on incorporating only high-quality methods, and tried to collate results omitting less reliable or less important ones.
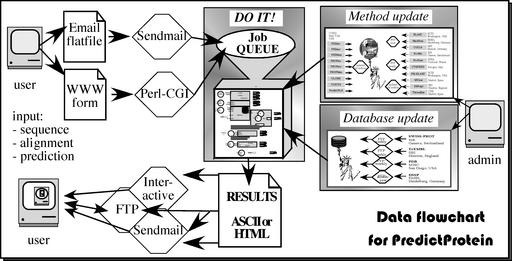
Figure 1.
Data flow for PredictProtein (PP). Users query PP with their protein and receive a single file with results from database comparisons and prediction methods. Users can query PP through email and web interfaces.
Figure 2.
Usage of PredictProtein. The number of requests to PP continues to rise (A); over one million requests came from 95 countries. Almost half of all the requests originated from North America (B).
Ultimate goal: collect all predictions for a protein in one image
Many of the servers launched after PP are either more specialised (e.g. only secondary structure prediction), or list all available tools irrespectively of performance accuracy. From the beginning, the ultimate goal of PP was to concatenate the results from all valuable and sustained prediction methods into one single image. This objective has driven us to build one of the most comprehensive public servers for sequence analysis and structure prediction. We hope to make the next leap into that direction in the near future.
Attempt to simplify output by incorporating hierarchy of thresholds
The attempt to ‘pre-digest’ as much information as possible to simplify the ease of interpreting the results is another unique pillar of PP. For example, by default PP returns only those proteins found in the database that are very likely to have a similar structure as the query protein (3). Particular predictions such as those for membrane helices, coiled-coil regions, signal peptides, nuclear localisation signals are not returned if found below given probability thresholds. Over the years, we have added so many methods into the output of PP that our original goal ‘easy-to-interpret’ is challenged. We hope that a variety of improvements in the near future will reduce this problem.
Each request triggers the application of over 10 different methods
Currently, users receive a single output file with the following results (some of these are optional, Table 1).
Database searches: similar sequences are reported and aligned by a standard, pairwise BLAST (4), an iterated PSI-BLAST search (5) and by the dynamic-programming method MaxHom (6). While the pairwise BLAST searches are identical to those obtainable from the NCBI site, the iterated PSI-BLAST is performed on a carefully filtered database to avoid accumulating false positives during the iteration (7,8). The dynamic-programming method MaxHom is only available through PP. Additionally database searches comprise a standard BLAST-based search through ProDom (9) and a standard search for functional motifs in the PROSITE database (10). Optionally, users can request searches for remotely similar proteins by the prediction-based threading method TOPITS (11).
Structure prediction methods: secondary structure, solvent accessibility, and membrane helices predicted by the PHD and PROF programs (12,13), coiled-coil regions by COILS (14) and bonded cysteine residues by CYSPRED (15). Putative structural switching regions are detected by the program ASP (16,17), low-complexity regions are marked by SEG (18) and long regions with no regular secondary structure are identified by NORSp (19). The PHD/PROF programs and TOPITS are only available through PP. The particular way in which PP automatically iterates PSI-BLAST searches and the way in which we decide what to include into sequence families is also unique to PP.
Table 1. Methods used by PP.
| Method | Task | Main author(s) | References |
|---|---|---|---|
| Database | |||
| SWISS-PROT* | Annotated protein sequences | A. Bairoch (SIB) and R. Apweiler (EBI) | (28) |
| TrEMBL* | Raw protein sequences | R. Apweiler (EBI) | (28) |
| PDB* | Protein structures | P. Bourne (UCSD) | (29) |
| BIG | Non-redundant combination of SWISS-PROT, TrEMBL,PDB | D. Przybylski (Columbia) | (7) |
| Alignment | |||
| MaxHom | Dynamic programming, multiple alignment | R. Schneider (LION) and C. Sander (Sloan Kettering) | (6) |
| BLASTP* | Pairwise alignment | S. Karlin and S. F. Altschul (NCBI) | (4) |
| PSI-BLAST* | Profile based alignment | S. F. Altschul (NCBI) | (5) |
| TOPITS | Prediction-based threading | B. Rost | (11,30,31) |
| Protein domains and unusual regions | |||
| ProDom* | Structural domain-like regions | F. Corpet, F. Servant, J. Gouzy and D. Kahn (Toulouse) | (32) |
| SEG* | Low-complexity regions | J. C. Wootton and S. Federhen (NCBI) | (18) |
| NORSp | Floppy regions | J. Liu and B. Rost | (19,33) |
| Protein structure | |||
| PHDsec | Secondary structure | B. Rost | (12,34,35) |
| PHDacc | Solvent accessibility | B. Rost | (12,36) |
| PHDhtm | Membrane helices | B. Rost | (12,37,38) |
| PROFsec | Secondary structure | B. Rost | (13) |
| PROFacc | Solvent accessibility | B. Rost | unpublished |
| GLOBE | Globularity | B. Rost | unpublished |
| COILS | Coiled-coiled regions | A. Lupas (Tübingen) | (39) |
| CYSPRED* | Disulfide-bonds | P. Fariselli and R. Casadio (Bologna) | (15) |
| ASP | Structural switches | M. Young and S. Highsmith (Sandia) | (17) |
| Protein function | |||
| PredictNLS | Nuclear localisation signals | R. Nair, M. Cokol and B. Rost (Columbia) | (40,41) |
| PROSITE* | Functional sequence motifs | K. Hofmann, P. Bucher and A. Bairoch (SIB) | (10) |
| Tools integrated into PP | |||
| Mview* | HTML alignment viewer | N. Brown | (42) |
| ESPript* | Ready-to-publish output for sequence alignments and secondary structure | P. Gouet and E. Courcelle (IPS Toulouse) | (43) |
*Original URLs are: SWISS-PROT, http://expasy.cbr.nrc.ca/sprot/; TrEMBL, http://www.ebi.ac.uk, PDB, http://www.rcsb.org/pdb/; BLASTP/PSI-BLAST, http://www.ncbi.nlm.nih.gov/BLAST/; PROSITE, http://www.expasy.ch/prosite/; ProDom, http://protein.toulouse.inra.fr/prodom.html; SEG, http://trex.musc.edu/manuals/unix/seg.html; CYSPRED, http://prion.biocomp.unibo.it/cyspred.html; MView, http://mathbio.nimr.mrc.ac.uk/~nbrown/mview/; ESPript, http://prodes.toulouse.inra.fr/ESPript.
Performance of methods
A detailed review about the strengths, weaknesses and pitfalls (20) of the many methods applied by PP is not possible here. Hence, we give only a brief overview over trends in the following.
Alignment methods: while the dynamic programming method MaxHom still appears best in aligning two proteins, the iterated PSI-BLAST tends to be more sensitive in unravelling more distantly related proteins. Note, however, that PSI-BLAST tends to over-estimate the relevance of short matches, and that PSI-BLAST expectation values have to be viewed with extreme caution when inferring similarity in function (21–23).
Protein domains and unusual regions: like, for instance, SMART (24), ProDom tends to identify regions that are significantly shorter than structural domains (25). Note that short regions of low complexity (SEG) are fairly common and not necessarily informative.
- Protein structure [see the EVA server (26) for an up-to-date evaluation of structure prediction]:
- 3a. PROFsec secondary structure prediction: on average, 76% of all residues are correctly predicted by (only ∼71% by PHDsec);
- 3b. PROFacc accessibility prediction: almost 80% of all residues are correctly predicted as either buried or exposed, and over 80% of the surface residues are correct;
- 3c. PHDhtm: ∼80% of the membrane helices are correctly predicted, for ∼66% of all tested proteins all membrane helices and the topology was correctly predicted (27); at the default threshold, membrane helices are incorrectly detected in ∼2% of the tested globular proteins (27); about one-fourth of all signal peptides (for secreted proteins) are mistaken for membrane helices (27);
- 3d. GLOBE: not accurate enough to identify domain boundaries, however, sufficient to capture trends like ‘very unlike a globular protein’;
- 3e. COILS: perceived to be correct most of the time;
- 3f. CYSPRED: most disulfide-bonding residues are correctly identified, however, most predicted bonds are wrong;
- 3g. ASP: if the protein has a structural switching region, this is usually detected correctly.
Note: the PROF and PHD series and CYSPRED are all based on artificial neural network systems.
INPUT, OUTPUT AND JOB OPTIONS
Default output
The output format is self-documenting. The output contains the following.
A list of likely homologues found in the protein database (BIG) and the multiple sequence alignment of these sequence (by default in ‘HTML’ format from MView).
If found: a list of the putative PROSITE motifs.
If found: a list of ProDom domain assignments.
If found: a prediction of coiled-coil regions.
Information about the expected levels of accuracy of structure predictions. (We suggest that newcomers read this carefully.)
- Prediction of aspects of protein structure. These are grouped in the following way:
- prediction of secondary structure for all residues;
- prediction of secondary structure for reliably scored residues only, with an expected three-state accuracy for these residues of >85%;
- prediction of solvent accessibility for all residues;
- prediction of solvent accessibility for reliably scored residues only, with an expected correlation between experimental observation and prediction of 0.69;
- prediction of transmembrane helices and their topology (if any detected).
Note: for the prediction of transmembrane helices a conservative threshold is chosen. Thus, a membrane helix may not be detected.
Advanced input options
By default users submit proteins through its one-letter residue sequence. However, PP also accepts submissions in FASTA, PIR and SWISS-PROT format or through the SWISS-PROT identifier. Most predictions methods applied use the information from the multiple alignments created by PP; prediction accuracy increases with the quality of the alignment. PP's alignments are fully automated, thus may not be as accurate as the alignment that experts have hand-edited. Therefore, users may also submit their favourite alignment directly. PP accepts alignments as FASTA lists, PIR lists, as well as in SAF and MSF format. The fold recognition/prediction-based threading method TOPITS uses predictions of secondary structure and solvent accessibility to search through a library of proteins of known structure. Predictions can be submitted through a simple column-based format.
Advanced prediction/job options
Not all methods are executed by default; some methods (like the prediction of membrane helices) use particular ‘conservative’ thresholds when included automatically and different thresholds when requested explicitly. In particular, the following methods can be toggled (switch on or off): MaxHom, BLASTP, PSI-BLAST, SEG, PHDsec, PHDacc, PHDhtm, PROFsec, PROFacc, COILS, CYSPRED, ASP, PROSITE, ProDom. Users can also explicitly request TOPITS or can evaluate the prediction accuracy of a secondary structure prediction method (EvalSec). Note that switching off methods has two advantages: it speeds up the execution and it reduces the size of the output. However, bear in mind that the database searches and their results are the limiting factor for speed and bytes produced.
Advanced output options
The default output now is an HTML formatted file, i.e. ready to display in any browser. Users can change this default to output in raw text in the following alignment formats: BLAST, no alignment, HSSP, HSSP profiles only, MSF, SAF, FASTA list. The results from the predictions are also available in a variety of machine-readable formats. (Developers: please do not write parsers for the human-readable PP output; in doubt, contact us, we can write almost any reasonable format if need be!) Due to the size of multiple alignments, we no longer email the results rather the output will be stored for a week on our web site (remember to download it in that period). Results can also be requested by email.
Interactive versus batch jobs
By default, the user submits requests to a batch queue and will be notified by email where to find the results (or will be sent these results). While PP also has an interactive mode that will write the results directly into the requesting web browser, this option comes with a restriction in the length of time for which the web connection is kept open: if PP has not completed a request within 5 min, we automatically switch the job to a batch mode and notify users by email. In practise, this implies that interactive jobs will only finish in time if (i) the PP queue is empty (works on a first-come-first-serve principle) and (ii) that the request does not require more than five minutes of CPU (typically the case if an alignment is submitted, and/or the query protein is short, and/or has few homologues in today's databases). We plan to upgrade the CPU resources for PP in the near future; this will increase the probability of successful interactive queries.
Acknowledgments
ACKNOWLEDGEMENTS
Making PredictProtein survive a decade was a major effort; many colleagues helped with hands and brains; thanks to all of them! Crucial contributions during the first years at EMBL: Antoine de Daruvar (Bordeaux University) wrote most of the initial server software, Reinhard Schneider (LION Biosciences, Heidelberg) helped with software and ideas, Sean O'Donoghue (LION Biosciences, Heidelberg) helped with encouraged continuation at difficult moments, Chris Sander (Sloan Kettering, New York) contributed his invaluable support, and Hans Doebeling and Björn Kindler provided sysadmin support. Thanks to Rolf Apweiler for his continued support at the European Bioinformatics Institute (EBI-EMBL, Hinxton, England), and to Volker Eyrich (Schrödinger, New York) for software support during the move to the USA. Further thanks to all who set up mirror pages and who consented to using their software in particular to Nigel Brown for MView, to Emmanuel Courcelle and Patrice Gouet (IPBS, Toulouse) for ESPript, to Florencio Pazos (Madrid) for Threadlize, to Henrik Nielsen and Søren Brunak (CBS, Copenhagen) for SignalP, to Andrei Lupas (Max Planck, Tübingen) for COILS, to Piero Fariselli and Rita Casadio (Bologna University) for CYSPRED, to Reinhard Schneider (LION Biosciences, Heidelberg) for MaxHom, to Malin Young (Sandia Labs, Albuquerque) for ASP, and to Rajesh Nair (Columbia University) for NLSpred. Finally, thanks to all the users who supported the service, in particular to Roland Walker (NCBI, Washington), Timothy Springer (Harvard, Cambridge, USA), Raphael Zidovetzki (UCI, Irvine), and Fernando Bazan (Stanford). Thanks to SGI, in particular to Juli Nash for crucial hardware support. J.L. and B.R. are supported by a grant to the Northeast Structural Genomics Consortium from the Protein Structure Initiative of National Institutes of Health (P50 GM62413). PredictProtein has attracted its first public support from the grant R01 LM07329-01 from the National Library of Medicine. Last, not least, thanks to Amos Bairoch (SIB, Geneva), Rolf Apweiler (EBI, Hinxton), Phil Bourne (San Diego University), and their crews for maintaining excellent databases and to all experimentalists who enable computational biology!
REFERENCES
- 1.Rost B. and Sander,C. (1992) Jury returns on structure prediction. Nature, 360, 540. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S. (1993) Sequence analysis by electronic mail server. Trends Biochem. Sci., 18, 267–268. [DOI] [PubMed] [Google Scholar]
- 3.Rost B. (1999) Twilight zone of protein sequence alignments. Protein Eng, 12, 85–94. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S.F. and Gish,W. (1996) Local alignment statistics. Methods Enzymol., 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S., Madden,T., Shaffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped BLAST and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sander C. and Schneider,R. (1991) Database of homology-derived structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 7.Przybylski D. and Rost,B. (2002) Alignments grow, secondary structure prediction improves. Proteins, 46, 195–205. [DOI] [PubMed] [Google Scholar]
- 8.Jones D.T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol., 292, 195–202. [DOI] [PubMed] [Google Scholar]
- 9.Corpet F., Gouzy,J. and Kahn,D. (1999) Recent improvements of the ProDom database of protein domain families. Nucleic Acids Res., 27, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rost B. (1995) TOPITS: Threading one-dimensional predictions into three-dimensional structures. In Rawlings,C., Clark,D., Altman,R., Hunter,L., Lengauer,T. and Wodak,S. (eds), Third International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp. 314–321. [PubMed] [Google Scholar]
- 12.Rost B. (1996) PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- 13.Rost B. (2001) Protein secondary structure prediction continues to rise. J. Struct. Biol., 134, 204–218. [DOI] [PubMed] [Google Scholar]
- 14.Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 15.Fariselli P., Riccobelli,P. and Casadio,R. (1999) Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins, 36, 340–346. [PubMed] [Google Scholar]
- 16.Kirshenbaum K., Young,M. and Highsmith,S. (1999) Predicting allosteric switches in myosins. Protein Sci., 8, 1806–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young M., Kirshenbaum,K., Dill,K.A. and Highsmith,S. (1999) Predicting conformational switches in proteins. Protein Sci., 8, 1752–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wootton J.C. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Tan,H. and Rost,B. (2002) Loopy proteins appear conserved in evolution. J. Mol. Biol., 322, 53–64. [DOI] [PubMed] [Google Scholar]
- 20.Rost B. and Valencia,A. (1996) Pitfalls of protein sequence analysis. Curr. Opin. Biotechnol., 7, 457–461. [DOI] [PubMed] [Google Scholar]
- 21.Rost B. (2002) Enzyme function less conserved than anticipated. J. Mol. Biol., 318, 595–608. [DOI] [PubMed] [Google Scholar]
- 22.Nair R. and Rost,B. (2002) Sequence conserved for sub-cellular localization. Protein Sci., 11, 2836–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devos D. and Valencia,A. (2001) Intrinsic errors in genome annotation. Trends Genet., 17, 429–431. [DOI] [PubMed] [Google Scholar]
- 24.Ponting C.P., Schultz,J., Milpetz,F. and Bork,P. (1999) SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res., 27, 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J. and Rost,B. (2003) Domains, motifs, and clusters in the protein universe. Curr. Opin. Chem. Biol., 7, 5–11. [DOI] [PubMed] [Google Scholar]
- 26.Koh I., Eyrich,V.A., Marti-Renom,M.A., Przybylski,D., Madhusudhan,M.S., Narayanan,E., Graña,O., Valencia,A., Sali,A. and Rost,B. (2003) EVA: evaluation of protein structure prediction servers. Nucleic Acids Res., 31, 3311–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C.P., Kernytsky,A. and Rost,B. (2002) Transmembrane helix predictions revisited. Protein Sci., 11, 2774–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman H.M., Westbrook,J., Feng,Z., Gillliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The protein data bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rost B., Schneider,R. and Sander,C. (1997) Protein fold recognition by prediction-based threading. J. Mol. Biol., 270, 471–480. [DOI] [PubMed] [Google Scholar]
- 31.Rost B. (1995) Fitting 1-D predictions into 3-D structures. In Bohr,H. and Brunak,S. (eds), Protein Folds: A Distance Based Approach. CRC Press, Boca Raton, Florida, pp. 132–151. [Google Scholar]
- 32.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J. and Rost,B. (2003) NORSp: predictions of long regions without regular secondary structure. Nucleic Acids Res., 31, 3833–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rost B. and Sander,C. (1993) Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- 35.Rost B. and Sander,C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins, 19, 55–72. [DOI] [PubMed] [Google Scholar]
- 36.Rost B. and Sander,C. (1994) Conservation and prediction of solvent accessibility in protein families. Proteins, 20, 216–226. [DOI] [PubMed] [Google Scholar]
- 37.Rost B., Casadio,R., Fariselli,P. and Sander,C. (1995) Prediction of helical transmembrane segments at 95% accuracy. Protein Sci., 4, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rost B., Casadio,R. and Fariselli,P. (1996) Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci., 5, 1704–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lupas A. (1996) Prediction and analysis of coiled-coil structures. Methods Enzymol., 266, 513–525. [DOI] [PubMed] [Google Scholar]
- 40.Cokol M., Nair,R. and Rost,B. (2000) Finding nuclear localisation signals. EMBO Reports, 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nair R., Carter,P. and Rost,B. (2003) NLSdb: database of nuclear localization signals. Nucleic Acids Res., 31, 397–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown N., Leroy,C. and Sander,C. (1998) MView: a web compatible database search or multiple alignment viewer. Bioinformatics, 14, 380–381. [DOI] [PubMed] [Google Scholar]
- 43.Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]