Abstract
Accurate T-cell epitope prediction is a principal objective of computational vaccinology. As a service to the immunology and vaccinology communities at large, we have implemented, as a server on the World Wide Web, a partial least squares-based multivariate statistical approach to the quantitative prediction of peptide binding to major histocom- patibility complexes (MHC), the key checkpoint on the antigen presentation pathway within adaptive cellular immunity. MHCPred implements robust statistical models for both Class I alleles (HLA-A*0101, HLA-A*0201, HLA-A*0202, HLA-A*0203, HLA-A*0206, HLA-A*0301, HLA-A*1101, HLA-A*3301, HLA-A*6801, HLA-A*6802 and HLA-B*3501) and Class II alleles (HLA-DRB*0401, HLA-DRB*0401 and HLA-DRB*0701). MHCPred is available from the URL: http://www.jenner.ac.uk/MHCPred.
INTRODUCTION
Vaccines induce protective immunity, an enhanced adaptive immune response to re-infection. In an era of failing antibiotics, vaccines, with their low cost and low frequency dosing, have generated renewed interest as prophylactic therapies. Historically, vaccines have been attenuated, or empirically weakened, whole pathogenic agents, such as viruses or bacteria. However, interest is now focusing on more rationally designed vaccines. These can be genetically modified pathogens, whole protein antigens or isolated poly-epitopes. Although the importance of non-peptide epitopes, such as lipids and carbohydrates, has become increasingly well recognized, it is the accurate prediction of proteinacious B-cell and T-cell epitopes (around which modern epitope-based vaccines are constructed) that remains the pivotal challenge for informatics with immunology. While B-cell epitope prediction remains unsophisticated (1), or is dependent on an often-indefinable knowledge of three-dimensional protein structure (2), a wide variety of advanced methods for T-cell epitopes prediction have arisen (3). It is generally accepted that only peptides that bind to major histocompatibility complexes (MHC) with an affinity above a threshold [typically a value of 500 nM (4)] function as T-cell epitopes and that peptide–MHC affinity roughly correlates with T-cell response. Most current methods for the prediction of T-cell epitopes depend on predicting peptides binding affinity to MHCs.
A few methods for MHC binding prediction have now been implemented as World Wide Web servers (Table 1). The provenance and utility of some of these servers remains uncertain, as their methods remain unpublished. In this paper, we present a noteworthy contribution to this field: a World Wide Web server, called MHCPred, which is a Perl implementation of our 2D QSAR approach to peptide–MHC prediction (5). MHCPred is available from the URL: http://www.jenner.ac.uk/MHCPred.
Table 1. Servers for peptide–MHC binding.
| Server name | Class | URL | Reference |
|---|---|---|---|
| SYFPEITHI | I and II | http://syfpeithi.bmi-heidelberg.com/Scripts/MHCServer.dll/EpiPredict.htm | 11 |
| BIMAS | I | http://bimas.dcrt.nih.gov/molbio/hla_bind/ | 13 |
| MHC-THREAD | II | http://www.csd.abdn.ac.uk/~gjlk/MHC-Thread/ | 18 |
| EpiPredict | II | http://www.epipredict.de/index.html | 19 |
| HLA-DR4 binding | II | http://www-dcs.nci.nih.gov/branches/surgery/sbprog.html | 20 |
| ProPred | II | http://www.imtech.res.in/raghava/propred/ | 21 |
| RankPep | I and II | http://www.mifoundation.org/Tools/rankpep.html | 22 |
| SVMHC | I | http://www.sbc.su.se/svmhc/ | 23 |
| PREDEP | I | http://bioinfo.md.huji.ac.il/marg/Teppred/mhc-bind/ | NP |
| NetMHC | I | http://www.cbs.dtu.dk/services/NetMHC/ | NP |
| PREDICT | I | http://sdmc.krdl.org.sg:8080/predict/ | NP |
| LpPep | I | http://reiner.bu.edu/zhiping/lppep.html | NP |
NP, not published.
SERVER DEVELOPMENT
Server software
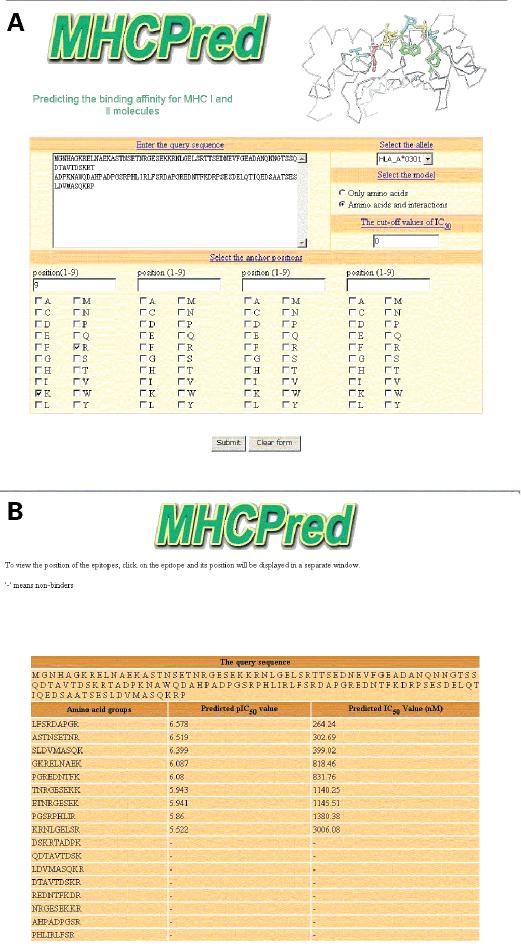
MHCPred runs as a CGI server, written in Perl, operating under Microsoft Windows NT. MHCPred is available from the URL: http://www.jenner.ac.uk/MHCPred. The interface is straightforward and intuitive: the sequence of a protein antigen is entered, an MHC allele and affinity threshold are selected and the program run (Fig. 1A). Additionally, an arbitrary motif can be entered to further restrain the search results. The results page produced subsequently displays a sorted list of nine amino acid substrings of the entered antigen sequence in order of calculated affinities (Fig. 1B).
Figure 1.
The MHCPred homepage and a prototypical results page. (A) The MHCPred homepage showing principal features of the interface: the sequence box, the choice of MHC allele model, the IC50 threshold, and the sets of checkboxes that allow a constraining sequence motif to be entered. A user defined motif, of 1–4 anchor positions, is entered as peptide positions plus corresponding allowed sets of amino acids: positions (1–9) are entered via a box and the allowed residues selected by clicking checkboxes corresponding to the required amino acids. Positions can be in any order but cannot be duplicated. (B) A MHCPred results page. A sorted list of subsequences and the calculated IC50 values are returned: each 9mer subsequence is shown, together with two columns: the −log IC50 and as the naked IC50 (units nM). Sequences scoring outside the threshold are shown as “−”. The output is constrained by the motif entered on the homepage.
MHCPred covers a range of different human MHC allele peptide specificity models. These include Class I (HLA-A*0101, HLA-A*0201, HLA-A*0202, HLA-A*0203, HLA-A*0206, HLA-A*0301, HLA-A*1101, HLA-A*3301, HLA-A*6801, HLA-A*6802 and HLA-B*3501) and Class II (HLA-DRB1*0101, HLA-DRB1*0401 and HLA-DRB1* 0701) alleles. All these alleles exist at a high frequency within human populations and have significant literature binding data.
Additive method
MHCPred models were generated from IC50 values obtained from radioligand competition assays characterizing peptide–MHC affinity. Values were collated from the literature and stored in the JenPep database (6). IC50 values were converted to log[1/IC50] values (or −log10[IC50] or pIC50) and are related to the free energy of binding: ΔGbind∼−RT ln IC50. pIC50 values were used as the dependent variables in a quantitative structure activity relationship (QSAR) regression. Generally, the dissociation constant varies with IC50, at least within one experiment, and, in practice, the variation in IC50 values is typically small enough such that values are comparable between experiments. The pIC50 values were predicted from a combination of individual amino acid contributions (P) at each position of the peptide and contributions from side chain–side chain interactions:
where the const accounts, at least nominally, for the peptide backbone contribution, ∑i=19Pi is the sum of amino acid contributions at each position and ∑j=18∑i=19−jPiPi+j is a series of summations for pairwise interactions between side chains of increasing sequence separation. In order to simplify this equation, we observe that class I MHC bound peptides assume extended but twisted conformations, so that adjacent side chains point in essentially opposite directions: both 1–2 and 1–3 interactions are possible between side chains. The resulting equation takes the form:
The need to handle data matrices with more variables than observations led us to use partial least squares (PLS) as our prediction engine and leave-one-out cross-validation to assess the predictive power of the models.
RESULTS
MHCPred is composed of a number of allele specific QSAR models created using PLS, a robust multivariate statistical method. Models of radioligand IC50 values, collated from the literature (6), were predicted using contributions from single amino acid side chains at each position and from interactions between 1–2 and 1–3 neighbours (5). Currently, MHCPred supports 11 class I HLA allele models and three Class II allele models.
Once a peptide has been bound by an MHC, for it to be recognized by the immune system the peptide–MHC complex has to be recognized by a T-cell receptor (TCR) of the T-cell repertoire. It is generally accepted that a peptide binding to an MHC may be recognized by a TCR if it binds better with a pIC50>6.3 or some similar figures for other binding measures (4). For this reason, MHCPred incorporates a user definable threshold for the pIC50 such that values below this are shown as non-binders. Generally, the approach taken is to whittle down the number of epitopes to a small number using a program such as MHCPred. These peptides are then tested as potential epitopes in T-cell activation assays.
In Table 2, we summarize quality measures for the PLS-based allele-specific multivariate statistical models we have generated. This range of models, which focuses primarily on the HLA-A locus, represents a set of alleles which are widely distributed in the human population and for which considerable binding data are available, leading to derived models with augmented predictivity. A useful result from the definition of MHC binding motifs, is that alleles can be grouped into so-called supertypes, which exhibit broad supermotifs, based on the commonality of their substrate specificity (7). Most of the models described above fall into the A2 and A3 supertypes. The model for A*0201 has been described in detail before (5) and details of other models will be published elsewhere (8–10). Table 3 compares MHCPred's efficiency in epitope prediction to that of the two best known methods: BIMAS (11) or SYFPEITHI (12).
Table 2. Evaluation of model statistics using PLS regression.
| n | q2 | NC | SEP | r2 | |
|---|---|---|---|---|---|
| A*0101 | 95 | 0.42 | 4 | 0.907 | 0.997 |
| A*0201 | 335 | 0.377 | 6 | 0.694 | 0.731 |
| A*0202 | 69 | 0.317 | 9 | 0.606 | 0.943 |
| A*0203 | 62 | 0.327 | 6 | 0.841 | 0.963 |
| A*0206 | 57 | 0.475 | 6 | 0.576 | 0.989 |
| A*0301 | 70 | 0.305 | 4 | 0.699 | 0.972 |
| A*1101 | 62 | 0.428 | 3 | 0.593 | 0.977 |
| A*3101 | 31 | 0.453 | 6 | 0.727 | 0.990 |
| A*6801 | 37 | 0.370 | 4 | 0.664 | 0.974 |
| A*6802 | 46 | 0.500 | 7 | 0.647 | 0.983 |
| B*3501 | 50 | 0.516 | 8 | 0.725 | 0.996 |
| DRB1*0101 | 90 | 0.808 | 8 | 0.567 | 0.994 |
| DRB1*0401 | 131 | 0.716 | 4 | 0.701 | 0.967 |
| DRB1*0701 | 84 | 0.649 | 7 | 0.562 | 0.999 |
n, Numbers of peptides; NC, number of components (others parameters as defined in 4).
Table 3. Comparison of MHCPred in T-cell epitope prediction.
| Protein | Protein | MHCPred | BIMAS | SYFPEITHI |
|---|---|---|---|---|
| H37Rv | Experimental T-cell epitopes found | 2 | 2 | 2 |
| Predicted binders | 6 | 13 | 9 | |
| HIV Vpr | Experimental T-cell epitopes found | 2 | 2 | 2 |
| HIV Vpr | Predicted binders | 6 | 6 | 6 |
| Experimental T-cell epitopes found | 2 | 2 | 2 | |
| Predicted binders | 5 | 14 | 6 |
Most experiments which identify T-cell epitopes use computational methods, however naive; typically this is the use of motifs to pre-select ‘high binders’ for testing. It is important, therefore, to use results only from experiments conducted with overlapping peptides covering the complete antigen sequence. Such comprehensive experiments are rare. Published results for three proteins are given (24,25): the number of experimentally observed epitopes are given together with the number of top ranked hits which contain them. MHCPred is at least as efficient as the other methods, and demonstrably more so in most cases.
Some of our models are more extensive than others, being built on larger more complete data sets, and have less absent values. As the number of absent values increases, the chance of incorrect prediction increases concomitantly. The coefficients for these missing values are set negative to decrease the number of false positive high binders predicted. As our database of peptide–MHC binding affinity measurements grows in size and diversity, we anticipate that absent value problems will ultimately disappear. In the meantime, we have implemented, within MHCPred, the option of imposing a sequence motif to limit the number of generated peptides. This allows for the informative combination of our quantitative, but probabilistic, models with other, essentially deterministic, motif models, which are available within SYFPEITHI (12), for example.
DISCUSSION
We have taken a quantitative approach to MHC binding prediction. This is important for several reasons. Firstly, binding affinity is the product of interactions of the whole peptide with a receptor molecule, and thus methods solely based on anchor positions (12) are likely to generate a significant proportion of both false-positive and false-negative predictions. Secondly, as our method is quantitative, it is directly applicable to the rational design of heteroclitic peptides (13), which are synthetically modified homologs of naturally occurring, mildly-immunogenic peptides that show increased MHC binding and augmented T-cell responses. Our approach allows the rapid identification of substitutions likely to increase binding and T-cell activation. Thirdly, it allows us a clear and unbiased comparison with experimental affinities. The only other quantitative method currently implemented on-line is BIMAS (11), which predicts the dissociation kinetics of the MHC-β2 microglobulin complex rather than the energetics of protein–ligand interaction as we do. Thus the two methods are complementary, although BIMAS is, unlike MHCPred, restricted solely to Class I.
Future developments of MHCPred will enhance both the scope and utility of the server and the method that underlies it. Firstly, we anticipate extending the number of allele models significantly, with an increased focus on both human class II MHCs and HLA-B and HLA-C loci, as well as non-human alleles, principally murine, bovine and primate. Although experimental data for peptide binding to class I alleles of lengths other than 9 is sparse, we will also seek to produce binding models of peptides of length 8, 10 and 11. We also envisage technical improvements directed towards automatic epitope mining of genomes. Likewise, by combining a user-defined set of allele models, we will be able to address the issue of identifying promiscuous peptides able to bind several different MHC alleles.
Secondly, the additive method implemented in MHCPred is, in itself, reliant upon the existence of particular amino acids at particular positions within peptides of the training set for it to predict reliably the effect of that residue at that position in any test peptides. Thus future technical developments will include the implementation of a descriptor-based, rather than an amino acid-based, approach to peptide QSAR, which will improve the generality of the method and allow us to model the effect of non-natural amino acids on binding (14,15).
CONCLUSION
The accurate prediction of T-cell epitopes is crucial to the development of computational vaccinology or computer aided vaccine design (16,17). The ability to predict MHC binding reliably will help us to analyze microbial immunomes, identifying the most antigenic epitopes and favoured putative vaccines. By making MHCPred available, we hope to foster collaboration within computer aided vaccine design. Such cooperation is vital if the field is to impact upon the discovery of novel vaccines in a way similar to that of other informatics techniques on the design, discovery and exploitation of new pharmaceuticals.
REFERENCES
- 1.Alix A.J. (1999) Predictive estimation of protein linear epitopes by using the program PEOPLE. Vaccine, 18, 311–314. [DOI] [PubMed] [Google Scholar]
- 2.Thornton J.M., Edwards,M.S., Taylor,W.R. and Barlow,D.J. (1986) Location of ‘continuous’ antigenic determinants in the protruding regions of proteins. EMBO J., 5, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flower D.R., Doytchinova,I.A., Paine,K., Taylor,P., Blythe,M.J., Lamponi,D., Zygouri,C., Guan,P., McSparron,H. and Kirkbride,H. (2002) Computational vaccine design. In Flower,D.R. (ed.), Drug Design: Cutting Edge Approaches. RSC Publications, London, pp. 136–180. [Google Scholar]
- 4.Sette A., Vitiello,A., Reherman,B., Fowler,P., Nayersina,R., Kast,W.M., Melief,C.J., Oseroff,C., Yuan,L. and Ruppert,J. (1994) The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol., 153, 5586–5592. [PubMed] [Google Scholar]
- 5.Doytchinova I.A., Blythe,M.J. and Flower,D.R. (2002) An additive method for the prediction of protein–peptide binding affinity. Application to the MHC class I molecule HLA-A*0201. J. Proteome Res., 1, 263–272. [DOI] [PubMed] [Google Scholar]
- 6.Blythe M.J., Doytchinova,I.A. and Flower,D.R. (2002) JenPep, a database of quantitative functional peptide data for immunology. Bioinformatics, 18, 434–439. [DOI] [PubMed] [Google Scholar]
- 7.Sinigaglia F. and Hammer,J. (1995) Motifs and supermotifs for MHC class II binding peptides. J. Exp. Med., 181, 449–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doytchinova I.A. and Flower,D.R. (2003) Towards the in silico identification of class II restricted T-cell epitopes: a partial least squares iterative self-consistent algorithm for affinity prediction. Bioinformatics, in press. [DOI] [PubMed] [Google Scholar]
- 9.Doytchinova I.A. and Flower,D.R. (2003) The HLA-A2 supermotif: a QSAR definition. Org. Biomol. Chem., in press. [DOI] [PubMed] [Google Scholar]
- 10.Guan P., Doytchinova,I.A. and Flower,D.R. (2003) HLA-A3-supermotif defined by quantitative structure-activity relationship analysis. Protein Eng., 16, 11–18. [DOI] [PubMed] [Google Scholar]
- 11.Parker K.C., Bednarek,M.A. and Coligan,J.E. (1994) Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol., 152, 163–175. [PubMed] [Google Scholar]
- 12.Rammensee H., Bachmann,J., Emmerich,N.P., Bachor,O.A. and Stevanovic,S. (1999) SYFPEITHI, a database for MHC ligands and peptide motifs. Immunogenetics, 50, 213–219. [DOI] [PubMed] [Google Scholar]
- 13.Tangri S., Ishioka,G.Y., Huang,X., Sidney,J., Southwood,S., Fikes,J. and Sette,A. (2001) Structural features of peptide analogs of human histocompatibility leukocyte antigen class I epitopes that are more potent and immunogenic than wild-type peptide. J. Exp. Med., 194, 833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellberg S., Sjostrom,M., Skagerberg,B. and Wold,S. (1987) Peptide quantitative structure–activity relationships, a multivariate approach. J. Med. Chem., 30, 1126–1135. [DOI] [PubMed] [Google Scholar]
- 15.Sandberg M., Eriksson,L., Jonsson,J., Sjostrom,M. and Wold,S. (1998) New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J. Med. Chem., 41, 2481–2491. [DOI] [PubMed] [Google Scholar]
- 16.Doytchinova I.A. and Flower,D.R. (2001) Toward the quantitative prediction of T-cell epitopes, CoMFA and CoMSIA studies of peptides with affinity for the class I MHC molecule HLA-A*0201. J. Med. Chem., 44, 3572–3581. [DOI] [PubMed] [Google Scholar]
- 17.Doytchiniva I.A. and Flower,D.R. (2002) Physicochemical explanation of peptide binding to HLA-A*0201 major histocompatibility complex. A three-dimensional quantitative structure–activity relationship study. Proteins, 48, 505–518. [DOI] [PubMed] [Google Scholar]
- 18.Swain M.T., Brooks,A.J. and Kemp,G.J.L. (2001) An automated approach to modelling class II MHC alleles and predicting peptide binding. Proceedings of the Second IEEE International Symposium on Bio-Informatics and Biomedical Engineering. IEEE Computer Society Press, pp. 81–88. [Google Scholar]
- 19.Fleckenstein B., Jung,G., von der Mulbe,F., Wessels,J., Niethammer,D. and Wiesmuller,K.H. (2001) From combinatorial libraries to MHC ligand motifs, T-cell superagonists and antagonists. Biologicals, 29, 179–181. [DOI] [PubMed] [Google Scholar]
- 20.Touloukian C.E., Leitner,W.W., Topalian,S.L., Li,Y.F., Robbins,P.F., Rosenberg,S.A. and Restifo,N.P. (2000) Identification of a MHC class II-restricted human gp100 epitope using DR4-IE transgenic mice. J. Immunol., 164, 3535–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh H. and Raghava,G.P. (2001) ProPred, prediction of HLA-DR binding sites. Bioinformatics, 17, 1236–1237. [DOI] [PubMed] [Google Scholar]
- 22.Reche P.A., Glutting,J.P. and Reinherz,E.L. (2002) Prediction of MHC class I binding peptides using profile motifs. Hum. Immunol., 63, 701–709. [DOI] [PubMed] [Google Scholar]
- 23.Donnes P. and Elofsson,A. (2002) Prediction of MHC class I binding peptides, using SVMHC. BMC Bioinformatics, 3, 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith S.M., Brookes,R., Klein,M.R., Malin,A.S., Lukey,P.T., King,A.S., Ogg,G.S., Hill,A.V. and Dockrell,H.M. (2000) Human CD8+ CTL specific for the mycobacterial major secreted antigen 85A. J. Immunol., 165, 7088–7095. [DOI] [PubMed] [Google Scholar]
- 25.Altfeld M.A., Livingston,B., Reshamwala,N., Nguyen,P.T., Addo,M.M., Shea,A., Newman,M., Fikes,J., Sidney,J., Wentworth,P. et al. (2001) Identification of novel HLA-A2-restricted human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte epitopes predicted by the HLA-A2 supertype peptide-binding motif. J. Virol., 75, 1301–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]