Abstract
Computed Atlas of Surface Topography of proteins (CASTp) provides an online resource for locating, delineating and measuring concave surface regions on three-dimensional structures of proteins. These include pockets located on protein surfaces and voids buried in the interior of proteins. The measurement includes the area and volume of pocket or void by solvent accessible surface model (Richards' surface) and by molecular surface model (Connolly's surface), all calculated analytically. CASTp can be used to study surface features and functional regions of proteins. CASTp includes a graphical user interface, flexible interactive visualization, as well as on-the-fly calculation for user uploaded structures. CASTp is updated daily and can be accessed at http://cast.engr.uic.edu.
INTRODUCTION
Protein performs its function through interaction with other molecules such as substrate, ligand, DNA and other domains of proteins. The three-dimensional structure of protein provides the necessary shape and physicochemical texture to facilitate these interactions. Structural information of protein surface regions enables detailed studies of the relationship of protein structure and function. Specifically, characterization of protein surface regions helps to analyze enzyme mechanism, to determine binding specificity and to plan mutation studies. It can also help to identify the biological roles of newly solved protein structures with an unknown function.
CASTp SERVER: VOIDS AND POCKETS OF PROTEINS
The CASTp web server aims to provide a comprehensive and detailed quantitative characterization of interior voids and surface pockets of proteins, which are prominent concave regions of proteins that are frequently associated with binding events (1,2). CASTp is based on the alpha shape and the pocket algorithm developed in computational geometry (2,3). In CASTp, voids are defined as buried unfilled empty space inside proteins after removing all hetero atoms that are inaccessible to water molecules (modeled as a spherical probe of 1.4 Å) from outside (4). Pockets are defined as concave caverns with constrictions at the opening on the surface regions of proteins. Unlike voids, pockets allow easy access of water probes from the outside.
CASTp identifies all pockets and voids on a protein structure and provides detailed delineation of all atoms participating in their formation. It also measures the volume and area of each pocket and void analytically, using both the solvent accessible surface model (Richards' surface) and molecular surface model (Connolly's surface). In addition, it measures the size of mouth openings of individual pockets, which helps to assess the accessibility of binding sites to various ligands and substrates. CASTp computation has been shown to be useful in a number of biological studies (5–10). The underlying algorithm and example of applications have been described elsewhere (2,3).
The CASTp server, in its most current release, was updated in January 2003. The previous version was launched in 1998 at the University of Minnesota. It is listed in the Research Consortium of Structural Bioinformatics PDB website at the San Diego Supercomputing Center (http://www.rcsb.org/pdb/), a central portal of structural bioinformatics worldwide. CASTp allows access to information of computed pockets and voids for structures in the Protein Data Bank (PDB). The server currently contains characterization of 1 322 538 pockets and voids that have been computed from 19 161 protein structures from the PDB. The CASTp server is updated nightly with new PDB entries.
USING CASTp
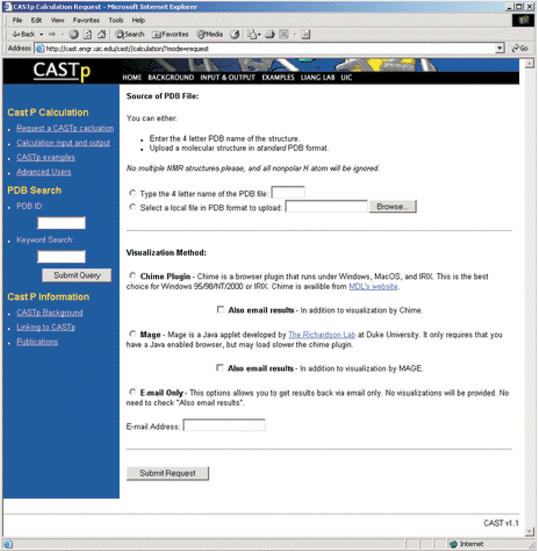
An intuitive graphic user interface allows querying of the CASTp server by typing the four letter PDB name of a protein structure, by keyword searching or by submitting their own molecular structure in the PDB format. Figure 1 shows the form for requesting a CASTp calculation. When querying by keyword, a list of relevant PDB structures are returned as obtained by redirecting to RCSB's PDB query site.
Figure 1.
An intuitive graphic user interface allows users to request a CASTp calculation by typing the four letter PDB name of a protein structure, by keyword searching or by submitting their own molecular structure in the PDB format. For visualization, the user has the choice of using MDL's Chime plug-in (Windows, MacOS and IRIX only) or Mage (11) java applet. In addition, users may also elect to have results emailed back as files.
For visualization, the user has the choice of using Chime plug-in developed by MDL Information Systems or Mage developed in the Richardson laboratory at Duke University (http://kinemage.biochem.duke.edu) (11). Chime is a browser plug-in that runs under Windows, MacOS and IRIX. This is a good choice for Windows 95/98/NT/2000 or IRIX. Mage is a Java applet that requires only a Java-enabled browser. This is a good choice for Linux platforms.
Figure 2 shows the binding site pocket on the catalytic domain of protein kinase A (pdb 2cpk) that is automatically identified by CASTp. Protein kinase A is an important enzyme in the signal transduction of vertebrates, participating in the regulation of glycogen synthesis, fatty acid synthesis, the oxidation of pyruvate to acteyl-CoA, mobilization of triacylglycerols and other processes. Many functionally important residues in the catalytic domain are located in this pocket, including the whole of the glycine rich loop (GTGSFGAV) starting from residue 50 that anchors ATP to the protein, the invariant K72 important to the optimum activity of the enzyme, the stabilizing E91 which forms a salt bridge with K72, residue E127 known to bind to the pseudosubstrate synthetic inhibitor, residue K168 in the catalytic loop which neutralizes the local negative charge of the gamma phosphate of the ATP, residue N171 which stabilizes the catalytic loop through hydrogen bonding to the backbone carbonyl of D166 (which is also identified) and residues D184 and G186 in the well-conserved triplet of DFG, where D184 is an invariant residue for orienting the gamma phosphate of MgATP for transfer to the substrate (12). Additional recent examples showing how CASTp computations have been used can be found in the literature (13–17).
Figure 2.
Visualization of ATP binding pocket automatically identified on the structure of the catalytic subunit of protein kinase A (pdb 2cpk). It is the largest pocket and contains the following residues: L49, G50, T51, G52, S53, F54, G55, R56, V57, A70, K72, L74, V79, N84, H87, T88, E91, L95, V104, M120, E121, Y122, V123, E127, D166, K168, E170, N171, L173, T183, D184, G186, F187, G200, T201 and F327. A large number of these residues are known to be functionally important (see http://www.sdsc.edu/kinases/) (12).
Once a protein is loaded in the browser, the user can interactively interrogate the protein structure. In addition to the simple manipulations built in to Chime plug-in and Mage applet (i.e. rotation, translation, zoom in/out), the user interface also allows selective highlighting of individual pockets. Summary information of measurement of individual pocket and void is conveniently displayed in a scrolling menu. Selection of a specific pocket from this menu also reveals the wall atoms comprising the pocket in a separate small window. By typing in the name and number of a residue, the user can easily identify the pocket or void that contains a particular residue.
Users may also choose the ‘Email only’ option to obtain results via email attachment. These include the following files: summary information of pockets and voids, summary information of mouth openings of pockets, atomic delineation of pocket walls, atomic delineation of mouth rims and a RasMol (18) and a kinemage (11) visualization script.
CASTp also supports on-the-fly calculations of voids and pockets on a structure uploaded by user. The user only needs to submit the coordinate file of a structure in the PDB format. When the calculation is finished, a visualization window will be opened. The results will also be emailed as an attachment to the user if requested. An added feature for the calculation of uploaded structures is that the user can adjust the probe radius to any values between 0.0 and 10.0 Å.
AVAILABILITY
CASTp can be freely accessed on the World Wide Web at http://cast.engr.uic.edu.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Herbert Edelsbrunner and Mike Facello for the alpha shape API library upon which the geometry engine of CASTp is implemented. We thank Dr Clare K. Woodward for encouragement. This work is supported by research grants from the National Science Foundation (CAREER DBI0133856 and DBI0078270).
REFERENCES
- 1.Lakowski R.A., Luscombe,N.M., Swindells,M.B. and Thornton,J.M. (1996) Protein clefts in molecular recognition and function. Protein Sci., 5, 2438–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang J., Edelsbrunner,H. and Woodward,C. (1998) Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci., 7, 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edeslbrunner H., Facello,M. and Liang,J. (1998) On the definition and the construction of pockets in macromolecules. Disc. Appl. Math., 88, 18–29. [PubMed] [Google Scholar]
- 4.Lee B. and Richards,F.M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol., 55, 379–400. [DOI] [PubMed] [Google Scholar]
- 5.Paetzel M. and Strynadka,N. (1999) Common protein architecture and binding sites in proteases utilizing a Ser/Lys dyad mechanism. Protein Sci., 8, 2533–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S., Liang,J. and Barry,B. (1997) Chemical complementation identifies a proton acceptor for redox-active tyrosin D in photosystem II. Proc. Natl Acad. Sci. USA, 94, 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LiCata V.J. and Bernlohr,D.A. (1998) Surface properties of adipocyte lipid-binding protein: Response to lipid binding, and comparison with homologous proteins. Proteins, 33, 577–589. [DOI] [PubMed] [Google Scholar]
- 8.Ory J.J. and Banaszak,L.J. (1999) Studies of the ligand binding reaction of adipocyte lipid binding protein using the fluorescent probe 1,8-anilinonaphthalene-8-sulfonate. Biophys. J., 77, 1107–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H., Raman,C., Glaser,C., Blasko,E., Young,T., Parkinson,J., Whitlow,M. and Poulos,T. (1999) Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. J. Biol. Chem., 274, 21276–21284. [DOI] [PubMed] [Google Scholar]
- 10.Thompson J., Reese-Wagoner,A. and Banaszak,L. (1999) Liver fatty acid binding protein: species variation and the accommodation of different ligands. Biochim. Biophys. Acta, 1441, 117–130. [DOI] [PubMed] [Google Scholar]
- 11.Richardson D.C. and Richardson,J.S. (1994) Kinemages-simple macromolecular graphics for interactive teaching and publication. Trends Biochem. Sci., 3, 135–138. [DOI] [PubMed] [Google Scholar]
- 12.Smith C., Shindyalov,I., Veretnik,S., Gribskov,M., Taylor,S., Ten Eyck,L. and Bourne,P. (1997) Development of Internet-based multimedia applications. Trends Biochem. Sci., 22, 444–446. [DOI] [PubMed] [Google Scholar]
- 13.Liu R., Pidikiti,R., Ha,C.E., Petersen,C.E., Bhagavan,N.V. and Eckenhoff,R.G. (2002) The role of electrostatic interactions in human serum albumin binding and stabilization by halothane. J. Biol. Chem., 277, 36373–36379. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J.R. and Banaszak,L.J. (2002) Lipid-protein interactions in lipovitellin. Biochemistry, 41, 9398–9409. [DOI] [PubMed] [Google Scholar]
- 15.Paetzel M., Dalbey,R.E. and Strynadka,N.C.J. (2002) Crystal structure of a bacterial signal peptidase apoenzyme—implications for signal peptide binding and the Ser-Lys dyad mechanism. J. Biol. Chem., 277, 9512–9519. [DOI] [PubMed] [Google Scholar]
- 16.Regni C., Tipton,P.A. and Beamer,L.J. (2002) Crystal structure of PMM/PGM: an enzyme in the biosynthetic pathway of P.aeruginosa virulence factors. Structure, 10, 269–279. [DOI] [PubMed] [Google Scholar]
- 17.Hilgers M.T. and Ludwig,M.L. (2001) Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site. Proc. Natl Acad. Sci. USA, 98, 11169–11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayle R.A. and Milner-White,E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]