Abstract
Karyotypic fission theory of Todd offers an explanation for the diverse range of diploid numbers of many mammalian taxa. Theoretically, a full complement of acrocentric chromosomes can be introduced into a population by chromosomal fission. Subsequent inheritance of ancestral chromosomes and paired fission derivatives potentially generates a diploid range from the ancestral condition to double its number of chromosomes. Although it is undisputed that both chromosomal fission and fusion (“Robertsonian rearrangements”) have significantly contributed to karyological diversity, it is generally assumed that independent events, the fission of single chromosomes or the fusion of two chromosomes, are the sources of such change. The karyotypic fission idea by contrast posits that all mediocentric chromosomes simultaneously fission. Here I propose a specific cell biological mechanism for Todd's karyotypic fission concept, “kinetochore reproduction theory,” where a complete set of dicentric chromatids is synthesized during gametogenesis, and kinetochore protein dephosphorylation regulates dicentric chromatid segregation. Three postulates of kinetochore reproduction theory are: (i) breakage of dicentric chromosomes between centromere pairs forms acrocentric derivatives, (ii) de novo capping of newly synthesized acrocentric ends with telomeric DNA stabilizes these derivatives, and (iii) mitotic checkpoints regulate chromosomal disjunction to generate fissioned karyotypes. Subsequent chromosomal rearrangement, especially pericentric inversion, increases the probability of genetic isolation amongst incipient sympatric species polytypic for fission-generated acrocentric autosomes. This mechanism obviates the requirement for numerous independent Robertsonian rearrangements and neatly accounts for mammalian karyotype evolution as exemplified in analyses of Carnivora, Artiodactyla, and Primates.
Karyotypic fission theory explains diverse mammalian karyotypes in taxa that include Canidae (2n = 34–78), Artiodactyla (2n = 14–74), Old World monkeys and apes (2n = 42–72), and lemurs (2n = 20–70) (1–4). The ancestral chromosomal complement for all mammals, in theory, was comprised of large mediocentric (i.e., metacentric, submetacentric, and subtelocentric) chromosomes (2n = 14), from which episodes of chromosome fission generated karyotypes with higher diploid numbers and smaller chromosomes (5). In populations with all mediocentric autosomes, a karyotypic fission event introduces a full complement of homologous acrocentric derivatives (1). Varying retention of ancestral mediocentric linkages and incorporation of fission-generated acrocentric pairs potentially generates a diploid range with up to twice the ancestral number of autosomes.
Karyotypic fission theory recently applied to lemurs (prosimian primates) explains their karyotypic diversity (2n = 20–70) with a minimum of four evolutionary steps, whereas prior explanations required at least 100 independent chromosomal mutations (4). The differential incorporation of fissioned chromosomes in five families (Lepilemuridae, Daubentoniidae, Lemuridae, Cheirogaleidae, and Indridae) accounts for extant karyotypes of all living lemur species. A fission event in the proposed ancestral karyotype for all lemurs (2n = 20) generated a diploid number range of 20–38. A secondary fission event in a karyologically polymorphic population (2n = 34, which either includes one acrocentric pair or is comprised of all mediocentric autosomes) explains all karyotypes of Lemuridae (2n = 44–62) and Cheirogaleidae (2n = 46 and 66), respectively. A separate secondary fission event in the ancestral stock of the Indridae (2n = 38, comprised of all mediocentric autosomes) explains all indrid karyotypes (2n = 40–70). A later independent fission event in the ancestral stock of the Lepilemuridae (2n = 20) explains their karyotypes (2n = 20–38). Karyotypic fission offers the most parsimonious explanation for the diverse chromosomal arrangements found in lemurs.
Karyotype Evolution in Animals.
Mammalian diploid numbers range from 6 to 92 (6, 7), and differences in chromosome number arise primarily through reorganization of chromatin by means of fission or fusion. Low diploid numbers (2n = 14–22) are found in didelphid marsupials, considered to be the most primitive true mammals (8). The homology between marsupial mediocentric chromosomes and smaller acrocentric pairs is shown by cross-species chromosome painting (9). Both Australian and South American marsupials exhibit a conserved complement of 2n = 14 with similar G-banding patterns (10). All higher marsupial diploid numbers can be explained by fission.
Karyological polymorphism is often best explained by fission theory, even in nonmammalian animal taxa. Chromosome morphology and DNA banding in Diprion pini and D. similis (Hymenoptera: Diprionidae) imply chromosome numbers doubled (from 2n = 14–28) by fission (11). The range of diploid numbers in the Australian ant genus Myrmecia (2n = 4–76) was interpreted as the result of fission (12). Fission accounts for karyotype diversity in reptiles; the iguana, Liolaemus monticola (2n = 38–44), and five species of colubrid snakes (2n = 28, 34, and 36) (13, 14). In the British Bay marine snail Nucella lapillus, typically monomorphic in diploid number (2n = 26), a few populations display variation in chromosome morphology (i.e., acrocentric pairs or mediocentric homologs) along a cline (2n = 26–36), which suggests fission as the best explanation. Trivalent formation during synapsis in N. lapillus hybrids involves 10 pairs of acrocentrics that correspond to 5 pairs of metacentric chromosomes (15). Homology between metacentric and subtelocentrics in N. lapillus hybrids was confirmed by DNA fluorescence in situ hybridization studies (16). That chromosomal diversity of such distinct taxa is explicable by fission implies this mode of animal evolution is important.
On the basis of recent advances in cell biology, I postulate “kinetochore reproduction theory” as the mechanism for the simultaneous fission of an entire chromosome complement. Biochemical behavior of kinetochores potentially leads to chromosome polymorphism and offers the most parsimonious explanation for fissioned chromosomes observed. I theorize that an additional round of kinetochore production followed by chromosomal breakage between kinetochore pairs simultaneously generates two acrocentrics for each mediocentric chromosome. A single plausible event: kinetochore reproduction that occurs during gametogenesis affects all autosomes. Consequent chromosomal rearrangement will produce normal offspring with no significant alteration of gene sequence, DNA quantity, or other phenotypic change.
Clinical studies and long-term mammalian cell cultures in which multiple dicentric chromosomes spontaneously arise provide evidence consistent with this theory (17). Transient multiple dicentric chromosomes were found in lymphocytes from populations in the United States, the United Kingdom, and Japan, and in South America Indians (18). The actual frequency of formation of multiple dicentric chromosomes is likely to be higher than that reported. Although details of supernumerary kinetochore development remain to be elucidated, the occurrence of this frequently detected viable and heritable chromosomal change is indisputable. My kinetochore reproduction theory attributes crucial significance to changes in the kinetochore reproduction timing that eventually lead to processes of species diversification in animal evolution.
Synchronous Reproduction of Kinetochores.
Kinetochores, disk-shaped proteinaceous structures, form at chromosome constrictions where sister chromatids join (i.e., on the surface of the centromere). These kinetochores, essential in spindle formation and chromosome segregation in eukaryotic cells (19, 20), are motors of chromatid segregation during karyokinesis (21). Kinetochore production during the interphase S stage when sister chromatids replicate is synchronized for an entire chromosome complement (22). Whereas a single kinetochore per centromere is seen during the G1 stage, typically the two kinetochores develop by late S stage and are visible in G2. I infer from the observation that kinetochores are synchronously produced that heritable changes generating additional kinetochore formation may affect all chromosomes within a cell. One additional round of kinetochore synthesis theoretically yields monocentric chromatids, each with a newly synthesized dicentric sister. Therefore, each chromosome has three rather than two active kinetochores. Inactivation of extra kinetochores involves the loss of kinetochore-associated protein (23); in the absence of such a loss, all kinetochores would be active during the next cell division.
Seeding New Kinetochores.
Two mechanisms of particular interest to this analysis for kinetochore synthesis are (i) the duplication of centromeric DNA that leads to neokinetochore formation, and (ii) epigenetic kinetochore formation in regions previously devoid of centromeric activity. Centromere proteins for kinetochore function are highly conserved (24), although centromeric DNA widely varies in sequence and quantity even among members of closely related species and races. The heterochromatin, the tandemly repeated α-satellite DNA associated with centromere/kinetochore formation, has been correlated with species divergence in great apes (25). Primate pericentromeric sequences “appear to have undergone unprecedented levels of duplication, transposition, inversion and either deletion or sequence divergence” in all species analyzed (26). That centromeric DNA evolves rapidly is significant to chromosomal evolution. The most parsimonious explanation for primate chromosomal evolution, the one that requires the least number of rearrangements, involves neocentromere formation (27). Mammalian kinetochores typically associate with specific centromeric DNA sequences. This association supports the hypothesis that simultaneous tandem duplication underlies the formation of supernumerary kinetochores. Duplication of centromeric DNA on all chromosomes results in a full complement of dicentric chromosomes. Kinetochores are known to form independently of standard centromeric sequences (28). Chromosomes within a karyotype even exhibit great variation in centromeric DNA, which suggests that the essential base pair sequence with an affinity for centromeric protein binding is minimal. Both duplication and epigenetic kinetochore formation provide likely basis for vertebrate chromosomal evolution by Todd's “karyotypic fission” theory.
Kinetochore Protein Dephosphorylation Regulates Chromosome Segregation.
During anaphase II, sister chromatids separate; those with duplicated centromeres must segregate from their monocentric sisters to ensure that each daughter cell receives a full complement of chromosomes (Fig. 1A). Chromosomal fission at this time results in partial aneuploidy. During division, a mechanoprotein-based “mitotic checkpoint” surveys kinetochore attachment. The onset of anaphase is postponed until all kinetochores properly attach to spindle. A transient delay in cell-cycle progression occurs while spindle attachment to chromosomes that have additional kinetochores is surveyed (29).
Figure 1.
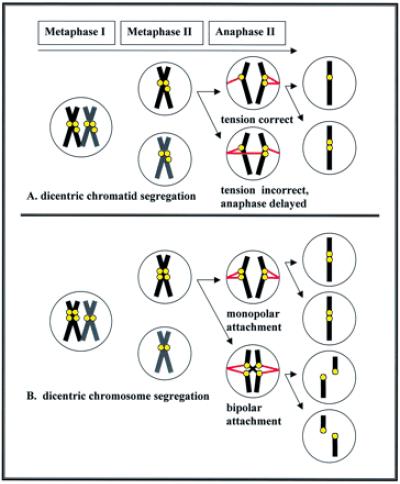
(A) Kinetochore production during gametogenesis generates dicentric chromatids. Surveillance of chromatid attachment delays anaphase until tension-sensitive kinetochore proteins respond to proper spindle attachment tension and signal resumption of cell cycle. Dicentric chromatids segregate from homologs and remain intact as they segregate. (B) Chromatids replicate, and fertilization leads to offspring heterozygotic for the dicentric chromosome. A mitotic delay occurs while chromatid attachment is surveyed at anaphase. When both kinetochores of a single chromatid attach facing the same pole in opposition to their homologs, chromosome segregates intact. Chromatids that have bipolar kinetochore attachments fission yielding two chromosomes, each with a functional kinetochore.
Kinetochores are active in the mitotic checkpoint mechanism. Checkpoint proteins at the kinetochore communicate with the “anaphase-promoting complex” to regulate the transition from metaphase to interphase. Key components of the mitotic checkpoint are Mad (mitotic arrest deficient) and Bub (budding uninhibited by benzimidazole) proteins first identified in yeast. In mammals and frogs, Mad2 localizes to unattached kinetochores during prometaphase and inhibits activation of the anaphase-promoting complex until all kinetochores have microtubule attachment (30). Chromatid separation is inhibited until proper alignment is achieved (31).
Tension-sensitive dephosphorylation of kinetochore proteins (putatively Bub1) provides a crucial signal for resumption of anaphase. In mammalian and insect cells, certain kinetochore proteins are phosphorylated before chromosome attachment to spindle (32). They dephosphorylate as soon as proper chromosome attachment occurs. During metaphase, the kinetochore proteins of chromosomes that are misattached remain phosphorylated, and anaphase does not ensue until kinetochore protein dephosphorylation occurs on all chromosomes. Kinetochore protein phosphorylation is effected by tension caused by spindle attachment. The Bub1 protein is postulated to be a tension-sensitive protein kinase that functions upstream of Mad2 to regulate the onset of anaphase in mammals (33). After dephosphorylation of Bub1, Mad2 disperses from kinetochores, and the anaphase-promoting complex is activated.
When the tension to kinetochores of the dicentric chromatid equals that of the kinetochore on its monocentric sister, proper segregation of dicentric chromatids in opposition to their monocentric sisters is assured. If a dicentric chromatid has monopolar attachment in opposition to its homolog, the tension will be correct. Whereas a single kinetochore on the dicentric chromatid pulled toward the same pole as the monocentric will have incorrect tension, dephosphorylation will not occur. Cell progression is delayed until proper alignment is complete.
Double-Strand Breakage Between Kinetochore Pairs.
Fertilization of a normal gamete with one that carries two kinetochores per chromatid will produce a heteromorphic hybrid. During the next round of DNA synthesis, dicentric chromatids become dicentric chromosomes with four kinetochores, two on each strand. Whether a dicentric chromosome breaks during anaphase depends on the orientation of kinetochore attachment to spindle (Fig. 1B). Breakage between kinetochores generates monocentrics in yeast (34, 35). If both chromatids have monopolar spindle attachment, they segregate intact and do not fission. However, if both chromatids have bipolar attachment in opposition to one another, double strand breaks will occur at weak points, which are likely to be at intercentromeric regions and have no deleterious effect on phenotype. Chromatin breakpoints are most frequently associated in the centromeric and telomeric regions at light G-bands areas (36). The de novo formation of stable fission-generated acrocentric derivatives of human chromosome 21 has no clinical significance, for example (37). Although fissioned autosomal homologs pair without difficulty, proper pairing of fissioned X and Y chromosomes may be inhibited by limited sex chromosome synapsis (at a small region of chromatin). Meiotic selection appears to favor the retention of ancestral nonfissioned sex chromosomes in mammals (1).
De Novo Telomeres.
Unless centromeric ends are promptly stabilized with telomeric DNA, unstable fissioned acrocentric derivatives may fuse with their counterparts or with other chromosomes or may form isochromosomes. Dicentric chromosomes in yeast form stable derivatives that exhibit newly acquired telomeric sequences (38, 39). Telomerase, a reverse transcriptase, maintains the distal sequences of chromosomes. Broken or missing telomeres are repaired by telomerase activity (40). Telomerase activity is notable in mammalian germline and embryonic cells (mice, ref. 41; humans, ref. 42) and also in oogenesis and embryogenesis of the amphibian Xenopus laevis (43). During a karyotypic fission event, the de novo acquisition of telomeric DNA on the centromeric ends of acrocentric derivatives is likely assured by high telomerase activity in embryonic cells.
Synapsis and Segregation of Acrocentric Derivatives.
During gametogenesis, homologous chromosomes randomly segregate. Inheritance of a pair of acrocentric derivatives or their homologous mediocentric chromosome is determined by chance. Two acrocentric derivatives synapse with their mediocentric homolog to form metaphase trivalents. DNA chromosome banding in hybrids confirms synapsis between homologs followed by normal disjunction of derivatives. Five homologous trivalents were observed in viable interspecific hybrid crosses between Eulemur fulvus fulvus (2n = 60) and Eulemur rubriventer (2n = 50) (44). Interspecific hybrid viability indicates that acrocentric pairs segregate together from their unfissioned homologs. If an acrocentric derivative begins to move toward the same pole as its mediocentric homolog, the tension on the two kinetochores is detected as incorrect, precluding dephosphorylation of kinetochore proteins. Anaphase will be delayed until proper tension is achieved. Trivalent synapsis was delayed in the mesquite lizard Sceloporus grammicus, heterozygotic for a fissioned chromosome (45). That these heteromorphic individuals suffer no meiotic deficit is consistent with my third postulate.
Homozygosity of fission-generated acrocentric pairs enables segregational independence for each. Homologous acrocentric chromosomes do synapse as typical bivalents that segregate independently from the other chromosomes of a complement. Although homozygosity for all chromosomes of a diploid is expected to be maximally stable, the many polymorphic states between a factor of two for chromosome number are sufficiently stable and viable to make kinetochore reproduction theory far more consistent with the enormous literature on animal karyotypes than the preconceived Robertsonian one-chromosome-change-at-a-time alternative.
Pericentric Inversions.
Pericentric inversion contributes to karyotypic polymorphism by relocation of the centromere, where an acrocentric chromosome is converted to a mediocentric or vice versa. Chromosome polymorphisms that were generated by pericentric inversion are common in mammals. A key postulate of Todd's karyotypic fission theory is the idea that in a postfissioned karyotype with a high number of acrocentric chromosomes, a trend for acrocentrics to revert to smaller mediocentrics by pericentric inversion (or centric fusion) repotentiates the karyotype for further fissions correlated with episodes of adaptive radiation directly inferable in the fossil record (46). Although karyological polymorphism that results from pericentric inversion generally does not reduce fertility, heterozygosity (inversion × no inversion) leads to difficulty during synapsis when large mediocentric chromosomes pair with smaller homologous pericentrically inverted derivatives. The gametic incompatibility accelerates genetic isolation in polymorphic species that, according to Todd's original thesis, augments sympatric speciation over time. Analysis of the fossil record of adaptive radiations for artiodactyls, carnivores, and primates is fully consistent with this view. Additional mammalian taxa that deserve scrutiny in light of karyotypic fission theory include insectivora, bats, and rodents.
Conclusions.
Karyotypic fission theory, backed by my kinetochore reproduction analysis, elegantly explains chromosomal diversification in mammals and probably other animal taxa. Kinetochore reproduction theory explains the cell biology that underlies specific chromosomal polymorphisms throughout the animal kingdom. The macroscopic analysis that posits the role of pericentric inversion and other chromosomal rearrangement in generating genetic incompatibility, reproductive isolation, and patterns of fossil taxa distribution enhances the generality of the theory. The fission model of evolution depicts radiations of diverse karyotypes supplemented by the accumulation of random mutations generally assumed, by themselves, to underlie speciation. Recent advances in cell-cycle regulation, chromosome behavior, fossil record, and phylogenetic inferences dispute that the primary direction of karyotypic evolution by sequential fusion of chromosomes is toward an arbitrary reduction in diploid number. Rather the tendency of kinetochores to reproduce, of telomerases to cap newly synthesized chromosome ends, and of mitotic checkpoints to regulate disjunction and generate freshly fissioned karyotypes in ancestral animals supports Todd's concept of saltatory chromosomal evolution. Increases up to nearly doubling of smaller derivative chromosomes throughout the Cenozoic underlie adaptive radiations, at least in artiodactyls, carnivores, lemurs, Old World monkeys, and apes.
Acknowledgments
I thank Dr. Lynn Margulis for instruction on kinetochores, Dr. Neil Todd for discussions about karyotypic fission, Drs. James Robl and Rafael Fissore for instruction in cell-cycle regulation, and Jeremy Sagan, Mark Kolnicki, and Michael Dolan for assistance in manuscript preparation.
References
- 1.Todd N B. J Theor Biol. 1970;26:445–480. doi: 10.1016/0022-5193(70)90096-2. [DOI] [PubMed] [Google Scholar]
- 2.Todd N B. Paleobiology. 1975;1:175–188. [Google Scholar]
- 3.Guisto J P, Margulis L. BioSystems. 1981;13:267–302. doi: 10.1016/0303-2647(81)90007-1. [DOI] [PubMed] [Google Scholar]
- 4.Kolnicki R L. Symbiosis. 1999;26:123–141. [Google Scholar]
- 5.Todd N B. Mammalian Chromosome Newsletter. 1967;8:268–279. [Google Scholar]
- 6.Hsu T C, Benirschke . An Atlas of Mammalian Chromosomes. 1–9. New York: Springer; 1967–1975. [Google Scholar]
- 7.Matthey R. In: The Chromosome Formulae of Eutherian Mammals, in Cytotaxonomy and Vertebrate Evolution. Chiarellia A B, Capanna E, editors. London: Academic; 1973. pp. 531–616. [Google Scholar]
- 8.Reig O A, Bianchi N O. Experientia. 1969;25:1210–1211. doi: 10.1007/BF01900283. [DOI] [PubMed] [Google Scholar]
- 9.O'Neill R J, Eldridge M D, Toder R, Ferguson-Smith M A, O'Brien P C, Graves J A. Genome. 1999;42:525–530. doi: 10.1139/g98-159. [DOI] [PubMed] [Google Scholar]
- 10.Rofe R, Hayman D. Cytogenet Cell Genet. 1985;39:40–50. doi: 10.1159/000132101. [DOI] [PubMed] [Google Scholar]
- 11.Rousselt J, Geri C, Hewitt G M, Lemuenier F. Heredity. 1998;81:573–578. doi: 10.1046/j.1365-2540.1998.00421.x. [DOI] [PubMed] [Google Scholar]
- 12.Hirai H, Yamamoto M T, Taylor R W, Imai H T. Chromosoma. 1996;105:190–196. doi: 10.1007/BF02509500. [DOI] [PubMed] [Google Scholar]
- 13.Lamborot M. Chromosome Res. 1998;6:247–254. doi: 10.1023/a:1009267821416. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez J M, Solorzano A, Cerdas L. Rev Biol Trop. 1984;32:263–267. [PubMed] [Google Scholar]
- 15.Bantock C R, Cockayne W C. Heredity. 1975;34:231–245. doi: 10.1038/hdy.1975.26. [DOI] [PubMed] [Google Scholar]
- 16.Pascoe P L, Patton S J, Critcher R, Dixon D R. Chromosoma. 1996;104:455–460. doi: 10.1007/BF00352269. [DOI] [PubMed] [Google Scholar]
- 17.Vig B K. Southest Asian J Trop Med Pub Health. 1995;26 Suppl. 1:68–76. [PubMed] [Google Scholar]
- 18.Awa A A, Neel J V. Proc Natl Acad Sci USA. 1986;83:1021–1025. doi: 10.1073/pnas.83.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbsky G J, Sammak P J, Borisy G G. J Cell Biol. 1987;104:9–18. doi: 10.1083/jcb.104.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorbsky G J, Sammak P J, Borisy G G. J Cell Biol. 1988;106:1185–1192. doi: 10.1083/jcb.106.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicklas B R. J Cell Biol. 1989;109:2234–2255. [Google Scholar]
- 22.Earnshaw W C, Tomkeil J E. Curr Opin Cell Biol. 1992;4:86–93. doi: 10.1016/0955-0674(92)90063-i. [DOI] [PubMed] [Google Scholar]
- 23.Earnshaw W C, Ratrie H, 3d, Stetten G. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- 24.Saffery R, Earle E, Irvine D V, Kalitis P, Choo K H. Chromosome Res. 1999;7:261–265. doi: 10.1023/a:1009222729850. [DOI] [PubMed] [Google Scholar]
- 25.Somonte R V, Ramesh K H, Verma R S. Genetica. 1997;101:97–104. doi: 10.1023/a:1018360026244. [DOI] [PubMed] [Google Scholar]
- 26.Jackson M S, Rocchi M, Thompson G, Hearn T, Crosier M, Guy J, Kirk D, Mulligan L, Rico A, Piccininni S, et al. Hum Mol Genet. 1999;8:205–215. doi: 10.1093/hmg/8.2.205. [DOI] [PubMed] [Google Scholar]
- 27.Montefalcone G, Tempesta S, Rocchi M, Archidiacono N. Genome Res. 1999;9:1184–1188. doi: 10.1101/gr.9.12.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saffery R, Irvine D V, Griffiths B, Kalitsis P, Wordeman L, Choo K, H. Hum Mol Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- 29.Yang S S, Yeh E, Salmon E D, Bloom K. J Cell Biol. 1997;136:345–354. doi: 10.1083/jcb.136.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waters J J, Chen R H, Murray A W, Gorbsky G J, Salmon E D, Nicklas R B. Curr Biol. 1999;9:649–652. doi: 10.1016/s0960-9822(99)80287-5. [DOI] [PubMed] [Google Scholar]
- 31.Skibbens R V, Hieter P. Annu Rev Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- 32.Nicklas R B, Campbell M S, Ward S C, Gorbsky G J. J Cell Sci. 1998;111:3189–3196. doi: 10.1242/jcs.111.21.3189. [DOI] [PubMed] [Google Scholar]
- 33.Taylor S S, McKeon F. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 34.Hill A, Bloom K. Mol Cell Biol. 1989;9:1368–1370. doi: 10.1128/mcb.9.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brock J K, Bloom K. J Cell Sci. 1994;107:891–902. doi: 10.1242/jcs.107.4.891. [DOI] [PubMed] [Google Scholar]
- 36.Yu C W, Borgaonkar D S, Bolling D R. Hum Hered. 1978;28:210–225. doi: 10.1159/000152960. [DOI] [PubMed] [Google Scholar]
- 37.Bogart M H, Fujita N, Serles L, Hsia Y E. Am J Med Genet. 1995;59:36–37. doi: 10.1002/ajmg.1320590108. [DOI] [PubMed] [Google Scholar]
- 38.Haber J E, Thorburn P C. Genetics. 1984;106:207–226. doi: 10.1093/genetics/106.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jager D, Phillipson P. EMBO J. 1989;8:247–254. doi: 10.1002/j.1460-2075.1989.tb03370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blackburn E H. Trends Biochem Sci. 1991;16:378–381. doi: 10.1016/0968-0004(91)90155-o. [DOI] [PubMed] [Google Scholar]
- 41.Prowse K R, Greider C W. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright W E, Piatyszek M A, Rainey W E, Byrd W, Shay J W. Dev Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 43.Mantell L L, Greider C W. EMBO J. 1994;13:3211–3217. doi: 10.1002/j.1460-2075.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rumpler Y, Dutrillaux B. Folia Primatol. 1980;33:253–261. doi: 10.1159/000155940. [DOI] [PubMed] [Google Scholar]
- 45.Reed K M, Sites J W, Greenbaum I F. Cytogenet Cell Genet. 1992;61:46–54. doi: 10.1159/000133367. [DOI] [PubMed] [Google Scholar]
- 46.Todd N B. In: Mammalian Evolution: Karyotypic Fission Theory, in Environmental Evolution. Margulis L, Olendzenski L, editors. Cambridge, MA: MIT Press; 1992. pp. 275–278. [Google Scholar]


