Abstract
The CRP (Cleavage of Radiolabeled Phosphoproteins) program guides the design and interpretation of experiments to identify protein phosphorylation sites by Edman sequencing of unseparated peptides. Traditionally, phosphorylation sites are determined by cleaving the phosphoprotein and separating the peptides for Edman 32P-phosphate release sequencing. CRP analysis of a phosphoprotein's sequence accelerates this process by omitting the separation step: given a protein sequence of interest, the CRP program performs an in silico proteolytic cleavage of the sequence and reports the predicted Edman cycles in which radioactivity would be observed if a given serine, threonine or tyrosine were phosphorylated. Experimentally observed cycles containing 32P can be compared with CRP predictions to confirm candidate sites and/or explore the ability of additional cleavage experiments to resolve remaining ambiguities. To reduce ambiguity, the phosphorylated residue (P-Tyr, P-Ser or P-Thr) can be determined experimentally, and CRP will ignore sites with alternative residues. CRP also provides simple predictions of likely phosphorylation sites using known kinase recognition motifs. The CRP interface is available at http://fasta.bioch.virginia.edu/crp.
INTRODUCTION
Functional proteomics is moving beyond the simple cataloging of protein content to describing the organization and functional state of proteins. The phosphorylation state of a protein is a critical determinant of function in signal transduction pathways, and is regulated by complex networks of kinases and phosphatases. Computational methods for predicting phosphorylation sites based on primary sequence lack both sensitivity and specificity (1); therefore, the phosphorylation state of a protein must continue to be measured experimentally. The traditional approach to phosphosite identification relies on the separation of proteolytic cleavage products and subsequent standard Edman protein sequencing; due to the need for peptide separation, this method is prohibitive for sensitive, high-throughput proteomics. A complementary, more sensitive method to rapidly identify phosphorylation sites uses simultaneous Edman phosphate release sequencing of unseparated 32P-labeled proteolytic cleavage products (2). Because no separation step is involved, the experiment can be performed quickly and is sensitive to femtomoles of starting material.
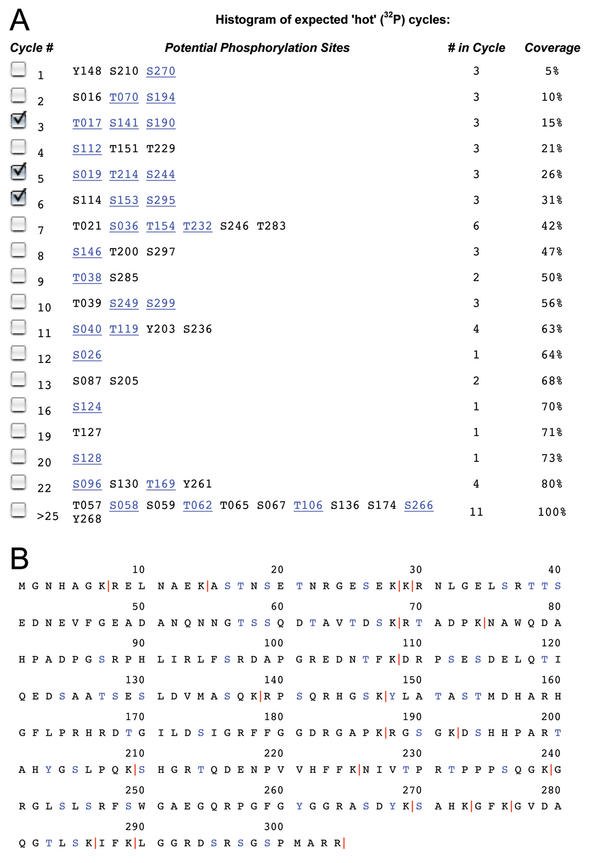
By simply observing the Edman cycles in which radioactivity is released and knowing the cleavage specificity of the proteolytic agent used to generate the peptides, candidate phosphorylation sites can be identified by their distance from the cleavage site; these distances will align with the radioactive Edman cycles. However, when two or more candidate phosphorylation sites are equally distant from a cleavage site the identification remains ambiguous: one or the other (or both) of the residues may be phosphorylated and be consistent with the observed data. For example, if the sequence of human myelin basic protein (MBP_HUMAN) is cleaved in silico at lysine (Fig. 1A), then Ser16, Thr70 and Ser194 would all be expected to appear in the second Edman cycle, as all three of these candidate phosphorylation sites are two residues away from cleavage sites at Lys12, Lys68 and Lys192 (Fig. 1B). Similarly, Thr17, Ser141 and Ser190 are three residues from lysines, and would appear in the third Edman cycle.
Figure 1.
(A) Prediction of candidate phosphorylation sites by CRP. The sequence for human myelin basic protein, a commonly used protein kinase substrate, was provided to CRP via its SWISS-PROT name ‘MBP_HUMAN’; cleavage at the carboxy-terminal of lysine was chosen. For cycles in which activity could be observed, a list of corresponding candidate phosphorylation sites is shown. The number in each cycle and the cumulative percent coverage is also provided. Candidate sites in the table that match known kinase specificities are hyperlinked to the corresponding PROSITE pattern record. Three cycles (3, 6 and >25) have been selected for further analysis; these would be selected because activity was observed in cycles 3 and 6 (see text). (B) The input sequence is provided for reference; red vertical bars reflect the cleavage site specifity, while candidate sites are blue.
On average, ambiguous assignments will occur in 80% of experiments (2). However, each experiment narrows the list of candidate phosphorylation sites to fewer residues, so that a subsequent experiment using a different proteolytic cleavage is more likely to resolve the ambiguity between the subset of candidates. Theoretically, over 70% of all known phosphorylation sites can be identified with two or three cleavage experiments (2). If an additional phosphoamino acid analysis is performed to identify the amino acid composition of the phosphorylated site(s), nearly 100% of known sites can be identified (2); for very long or hyperphosphorylated proteins, phosphoamino acid analyses may be required to obtain meaningful results. The CRP program guides this experimental design and helps interpret the results.
DESCRIPTION
The CRP program is a Perl CGI-based WWW script that performs an in silico sequence digestion, counts the number of residues (or Edman cycles) between each serine, threonine or tyrosine and the cleavage site, and tabulates the theoretical results. The program accepts: (i) either a protein sequence (in FASTA or raw sequence format) or a unique Entrez identifier (e.g. a GenBank GI number, accession number or SWISS-PROT name); (ii) the choice of proteolytic reagent to be used (commonly available endoproteinases and their cleavage patterns are listed, but the user may provide an alternative pattern); (iii) whether the reagent exhibits prolyl-resistant cleavage; (iv) a list of phosphoamino acids to consider (by default, all Ser, Thr and Tyr are included in the analysis); and (v) a cycle cutoff above which positions cannot be experimentally resolved. Once submitted, CRP calculates a histogram of possible Edman cycles in which radioactivity might be observed (Fig. 1A), including the candidate phosphorylation sites that are associated with radioactivity in each cycle. The histogram table also indicates the cumulative candidate site coverage at each cycle number. Candidate sites that match known kinase substrate specifities are highlighted and hyperlinked to their corresponding PROSITE record (3). The input sequence illustrates the positions of cleavage sites (vertical red bars) and candidate phosphorylation sites (colored blue) (Fig. 1B). Experimental results are compared with those predicted by the CRP program: if only one candidate site is found in the cycle(s) exhibiting radioactivity, then identification is complete and unambiguous.
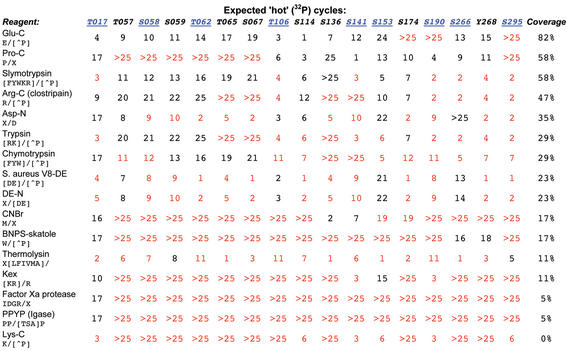
When an unambiguous identification cannot be made, CRP can be used to plan and/or interpret a second cleavage experiment to resolve the ambiguity. All cycles in the first experiment that exhibit radioactivity (including any that appear at higher cycle numbers than experimentally measured) should be marked for further processing. CRP tabulates a new set of cycle numbers where each of these candidates would appear, with varying cleavage specifity (Fig. 2), ranking reagents by their ability to uniquely identify the remaining sites. These tables can help identify a strategy for further experimentation, suggesting cleavage sites that resolve the ambiguity within experimentally achievable cycle numbers (usually limited to 30–40 cycles).
Figure 2.
Theoretical cleavage results for human MBP, focusing on the candidate sites selected in Figure 1. For each candidate site, predicted radioactive cycle numbers are listed by cleavage reagent. Red cycle numbers indicate that identification of this site would remain ambiguous with the given reagent. The table shows that a second experiment using endoproteinase Glu-C would allow 82% of the candidate sites to be unambiguously identified.
DISCUSSION
Experimental measurement remains the only reliable method to ascertain a protein's phosphorylation state. Prediction of candidate phosphorylation sites through sequence motifs (3,4) suffers from unacceptably low sensitivity; more sophisticated machine-learning algorithms improve sensitivity in exchange for poor selectivity (1). Structurally-weighted sequence motifs may provide better performance (5). Moreover, none of these methods can predict changes in phosphorylation with time or cell type.
Traditional Edman protein sequencing approaches are fast being replaced by more sensitive and much faster mass spectrometry (MS)-based methods for proteomic analyses, capable of identifying the phosphorylation state of a protein (6,7). However, MS phosphoprotein experiments are expensive, and achieve limited sensitivity at low sample concentrations. Alternatively, CRP-assisted Edman phosphate-release sequencing of unseparated radiolabelled phosphopeptides can achieve high sensitivity due to the lack of chromatographic sample loss, but requires a somewhat more sophisticated analysis than traditional Edman sequencing to interpret the data. Additional experimentation may also be required to resolve ambiguities. As an experimental planning tool, CRP aids in the choice of cleavage site to achieve maximal unambiguous coverage; in a high-throughput proteomic setting, multiple cleavage experiments could be performed in parallel, and CRP would help identify unambiguous phosphorylation sites.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to our reviewers for their helpful suggestions. A.J.M. and W.R.P. are supported by Grant LM04961 from the National Library of Medicine; T.A.J.H. is supported by Grants HL19242-24 and DK52378-01 from the National Institutes of Health.
REFERENCES
- 1.Blom N., Gammeltoft,S. and Brunak,S. (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol., 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald J.A., Mackey,A.J., Pearson,W.R. and Haystead,T.A. (2002) A strategy for the rapid identification of phosphorylation sites in the phosphoproteome. Mol. Cell Proteom., 1, 314–322. [DOI] [PubMed] [Google Scholar]
- 3.Sigrist C.J., Cerutti,L., Hulo,N., Gattiker,A., Falquet,L., Pagni,M., Bairoch,A. and Bucher,P. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform., 3, 265–274. [DOI] [PubMed] [Google Scholar]
- 4.Gattiker A., Gasteiger,E. and Bairoch,A. (2002) ScanProsite: a reference implementation of a PROSITE scanning tool. App. Bioinform., 1, 107–108. [PubMed] [Google Scholar]
- 5.Brinkworth R.I., Breinl,R.A. and Kobe,B. (2003) Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc. Natl Acad. Sci. USA, 100, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H., Watts,J.D. and Aebersold,R. (2001) A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol., 19, 375–378. [DOI] [PubMed] [Google Scholar]
- 7.Steen H., Kuster,B., Fernandez,M., Pandey,A. and Mann,M. (2002) Tyrosine phosphorylation mapping of the epidermal growth factor receptor signaling pathway. J. Biol. Chem., 277, 1031–1039. [DOI] [PubMed] [Google Scholar]