Abstract
LOC3D (http://cubic.bioc.columbia.edu/db/LOC3d/) is both a weekly-updated database and a web server for predictions of sub-cellular localization for eukaryotic proteins of known three-dimensional (3D) structure. Localization is predicted using four different methods: (i) PredictNLS, prediction of nuclear proteins through nuclear localization signals; (ii) LOChom, inferring localization through sequence homology; (iii) LOCkey, inferring localization through automatic text analysis of SWISS-PROT keywords; and (iv) LOC3Dini, ab initio prediction through a system of neural networks and vector support machines. The final prediction is based on the method that predicts localization with the highest confidence. The LOC3D database currently contains predictions for >8700 eukaryotic protein chains taken from the Protein Data Bank (PDB). The web server can be used to predict sub-cellular localization for proteins for which only a predicted structure is available from threading servers. This makes the resource of particular interest to structural genomics initiatives.
INTRODUCTION
Sub-cellular localization is one aspect of protein function
Proteins must be localized in the same sub-cellular compartment to co-operate towards a common biological function. Thus, the native sub-cellular localization of a protein is important for the understanding of gene/protein function. Aberrant sub-cellular localization of proteins has been observed in the cells of several diseases, such as cancer and Alzheimer's disease. Therefore, experimentally unravelling the native compartment of a protein constitutes one step on the long way to determining its function. The explosion of sequence information through large-scale sequencing projects has widened the gap between the number of sequences deposited in public databases and the experimental characterization of the corresponding proteins (1,2). Experimental annotations of sub-cellular localization are often based on operational, biochemical definitions (e.g. cell fractions or targeting signals of various sorts) that can be error prone. In contrast, computational tools can provide fast and accurate localization predictions for any organism (3,4). Attempts to predict sub-cellular localization have become one of the central problems in bioinformatics (5,6).
Predicting sub-cellular localization of proteins
The Protein Data Bank (PDB) (7) contains proteins of known three dimensional (3D) structures. Sub-cellular localization is annotated for very few of the proteins deposited in PDB. The LOC3Ddb database is the first comprehensive database of predicted and inferred sub-cellular localization for proteins of known structure. Four different methods are applied (see Methods); the method with the strongest signal is chosen to annotate sub-cellular localization for all proteins in PDB. The LOC3Ddb database can be useful in complementing functional information for proteins from domain databases like SMART (8), PFAM (9) and functional site resources like ELM (10), ProtFun (11) and Prosite (12).
METHODS
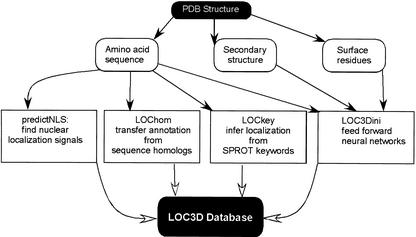
LOC3D ventures four different paths to annotate sub-cellular localization (Fig. 1), these are: (i) PredictNLS; (ii) LOChom; (iii) LOCkey; and (iv) LOC3Dini.
Figure 1.
The LOC3D system. From the query PDB structure, the amino acid sequence, three state secondary structure and solvent accessible surface residues of the protein are extracted. LOC3D uses four different methods to annotate sub-cellular localization as follows. (i) PredictNLS: the amino acid sequence is scanned for nuclear localization signals. (ii) LOChom: the sequence is first aligned using PSI-BLAST to a localization annotated database of proteins. If any sequence homologues are discovered, sub-cellular localization annotation is transferred from the homologue. (iii) LOCkey: the SWISS-PROT database contains functional information for proteins in the form of keywords. LOCkey infers sub-cellular localization based on keyword entries. The above three programs are based solely on the amino acid sequence of the protein and do not use any structural information. (iv) LOC3Dini: sub-cellular localization is predicted by a system of neural networks trained on a number of global features like amino acid composition, secondary structure composition and surface residue composition. The final localization annotation in the LOC3D database is taken from the most reliable prediction amongst the four individual methods.
PredictNLS: identification of nuclear localization signals
The most accurate way to predict nuclear localization is to identify the nuclear localization signal: active transport of proteins into the nucleus takes place by binding to specific molecules such as importins and karyopherins that recognize distinct targeting signals (13). This targeting signal typically contains a short segment of consecutive residues and is commonly referred to as the nuclear localization signal (NLS). PredictNLS (14,15) uses a set of expert-curated experimentally known NLSs to predict nuclear localization. At 100% accuracy this tool identifies ∼50% of all known nuclear proteins.
LOCkey: digest experimental data from SWISS-PROT keywords
Our second most accurate tool to infer localization is to simply infer localization from experimental descriptions of localization as contained in the controlled vocabulary of SWISS-PROT (16). LOCkey infers sub-cellular localization through an automated lexical analysis of SWISS-PROT keywords (17). In contrast to dictionary-based approaches, LOCkey is fully automated and the rule libraries used to infer localization from keywords are generated dynamically. The method is based on a novel implementation of an M-ary (multiple category) classifier (18,19). For example for a protein with the SWISS-PROT keywords ‘NADP, Acetylation, NAD and Oxidoreductase’ (keywords for the protein and its sequence homologs are merged to obtain the final keyword list), the LOCkey algorithm first generates all possible combinations of the keywords. For a protein with four keywords this gives 24−1=15 possible combinations (examples of keyword combinations are ‘NADP, Acetylation’, ‘Acetylation, NAD, Oxidoreductase’, ‘Acetylation, NAD’, ‘NADP’ and so on). For each of these keyword combinations the algorithm compiles localization statistics for matches against a pre-compiled database of keyword associations for proteins of known sub-cellular localization. Finally localization is assigned to the protein by minimizing an entropy-based objective function (in this example the localization assigned is Cytoplasm). The method is extremely accurate in inferring sub-cellular localization when any functional information in the form of keywords is known (>82% accuracy using full cross-validation).
LOChom: inference through sequence homology
The next most reliable means of getting at sub-cellular localization is through annotation transfer through homology: if a protein of experimentally known localization L is significantly similar in sequence to a query protein U, U and L have identical localization (20,21). We have carried out the most exhaustive study of the sequence conservation of sub-cellular localization to establish the thresholds for annotation transfer based on homology (20). Sequence homologs were identified using pairwise BLAST (22) and PSI-BLAST (23). To assign sub-cellular localization three measures of sequence similarity were investigated: (i) sequence identity; (ii) BLAST e-values (22); and (iii) distance from ‘HSSP-threshold’ (24,25). Of the three measures, distance from ‘HSSP-threshold’ was the most successful in annotating sub-cellular localization. One of the results of this investigation was a refined version of the distance from ‘HSSP-threshold’ formula (20,25,26) which significantly improves the coverage of proteins that can be annotated using homology. The use of position specific scoring matrix (PSSM) in PSI-BLAST improved the coverage of proteins that could be annotated using homology by >5% over simple pairwise BLAST. Further improvements in homology-based annotation were obtained through the use of separate ‘conservation thresholds’ and ‘accuracy versus sequence similarity’ curves for each of the localization classes.
LOC3Dini: ab initio prediction from sequence and structure
LOC3Dini is a prediction system that predicts sub-cellular localization from sequence and structure using neural networks (manuscript submitted). Sub-cellular localization is predicted using a number of global features of protein sequence and structure. The LOC3Dini system consists of three layers and sorts proteins into one of four localization classes (extracellular, cytoplasmic, nuclear and mitochondrial).
The first layer consists of four dedicated neural networks that use particular features from protein sequences, alignments and secondary structure to pre-sort proteins into L/not-L (with L=cytoplasmic, nuclear, extra-cellular or mitochondrial). The features used include, amino acid composition, composition of surface accessible residues and composition of amino acid residues in one of three secondary structure states (helix, beta strand and loop). Evolutionary information was incorporated by replacing the amino acid composition by profile based amino acid composition.
The second layer consists of neural networks combining output from networks trained on different input features.
The third layer uses a simple jury decision to assign one of four localization states to each protein.
Major sources of improvement over publicly available methods originated from using: (i) secondary structure information; (ii) solvent accessibility; and (iii) evolutionary information from sequence profiles as input to the neural networks. The final four-state classification accuracy of the system was >65%. This is >10 percentage points higher than systems using only amino acid composition.
Final annotation of sub-cellular localization through best single method
The final annotation of localization that is generated by LOC3D is taken from the most reliable prediction amongst the four individual methods. Using this four-step approach significantly improves prediction accuracy since different methods are most accurate in different regimes. For example, if an NLS is detected by PredictNLS, the protein has a high probability of being nuclear (our NLS motifs are exclusive to nuclear proteins). If functional information in the form of SWISS-PROT keywords is available, LOCkey can use this information to infer sub-cellular localization at a very high accuracy. In the absence of sufficient functional information, identification of sequence homologs using LOChom proves most accurate. Ab initio predictions using LOC3Dini are the least accurate of the four methods; however, they are applicable when all the other methods fail. In terms of coverage of PDB, the most successful methods were homology-based assignments using LOChom, accounting for 44% of the final assignments and keywords-based assignments using LOCkey, accounting for 37% of the assignments (Table 1). Although extremely accurate, nuclear localization signals could be inferred for only ∼1% of the eukaryotic chains using PredictNLS.
Table 1. Annotations by LOC3D by method.
| Method | Number of proteinsa | Percentage of proteins |
|---|---|---|
| LOChom | 3880 | 44 |
| LOCkey | 3222 | 37 |
| LOC3Dini | 1561 | 18 |
| PredictNLS | 130 | 1 |
| SUM | 8793 | 100 |
aNumber of final localization assignments madeby each method.
Comprehensive annotation for 3D structures
LOC3D is a comprehensive source of information regarding sub-cellular localization for eukaryotic proteins of known structure. Currently, there are no available databases cataloguing sub-cellular localization information for eukaryotic chains in PDB. The database contains sub-cellular localization information for >8700 PDB chains (Table 2). Proteins secreted to the extra-cellular space form the largest class of proteins in PDB, followed by cytoplasmic and nuclear proteins.
Table 2. Annotations by LOC3D by type of localization.
| Sub-cellular localization | Number of proteinsa |
|---|---|
| Extracellular space | 3786 |
| Cytoplasm | 2328 |
| Nucleus | 1066 |
| Mitochondria | 1024 |
| Chloroplast | 348 |
| Peroxysome | 88 |
| Lysosome | 85 |
| Endoplasmic reticulum | 50 |
| Vacuoles | 14 |
| Golgi apparatus | 4 |
| Total | 8793 |
aNumber of PDB chains in the LOC3D detabase assigned to the given localization.
LOC3D INTERFACE
Database description
The LOC3D database has been formatted in an EMBL-like flat-file format. The database can be accessed on the web through a PERL CGI interface (27–29). The database can be used in either query mode or browse mode.
User query: any object in the database can be queried using a PERL regular expression like syntax. The query can be a name or a wildcard pattern (the search engine automatically appends the ‘*’ wildcard pattern at the end of the query). If the query field is left blank, the search displays all objects of the selected type. Three types of objects can be queried: PDB chain identifiers, types of sub-cellular localization and type of prediction method. For example, querying the ‘sub-cellular localization class’ object with ‘nuclear’ displays all proteins in the database that are predicted to have nuclear localization.
Browsing the database: in this mode, database entries are displayed in order of decreasing confidence of prediction.
Web server description
The LOC3D web server has been implemented using a PERL CGI interface. Protein structures in PDB format (7) can be submitted to the server. Sub-cellular localization is predicted using the method described above. Prediction results are returned via email.
Format and fields
Each protein can have up to four localization predictions associated with it, one from each method. The database uses four fields to represent predictions from each method:
Method: the type of prediction method used.
Loci: predicted sub-cellular localization from this method. The predicted sub-cellular localization can be one of nine classes (Table 2).
Confidence: confidence score assigned by the prediction method. This is a number between 0 and 100. Larger confidence scores mark more accurate predictions.
Details: any reasons, if available, for the particular localization class inferred by the method. For example, for a LOCkey prediction, this field would give details of the keywords responsible for this localization prediction.
CONCLUSIONS
LOC3D should be a useful resource for functional studies of proteins. In particular, large-scale efforts in structural genomics may profit from the tool. We plan to implement the system to predict sub-cellular localization based on predicted secondary structure. Another future goal is to also include all proteins in the SWISS-PROT database and the fully-sequenced eukaryotic genomes. We also plan to incorporate the database into comprehensive proteome databases like the PEP (30) database.
LOC3D should be cited with the present publication as reference. The database can be accessed through the World Wide Web at: http://cubic.bioc.columbia.edu/db/LOC3D/.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Jinfeng Liu and Megan Restuccia (Columbia) for computer assistance and Kaz (Columbia) for valuable discussions. The work of R.N. and B.R. was supported by grant DBI-0131168 from the National Science Foundation (NSF). Last, but not least, we are grateful to Amos Bairoch (SIB, Geneva), Rolf Apweiler (EBI, Hinxton), Phil Bourne (San Diego University) and their crews for maintaining excellent databases and to all experimentalists who enabled this tool by making their data publicly available.
REFERENCES
- 1.Koonin E.V. (2000) Bridging the gap between sequence and function. Trends Genet., 16, 16. [DOI] [PubMed] [Google Scholar]
- 2.Rost B. and Sander,C. (1996) Bridging the protein sequence-structure gap by structure predictions. Ann. Rev. Biophys. Biomol. Struct., 25, 113–136. [DOI] [PubMed] [Google Scholar]
- 3.Lewis S., Ashburner,M. and Reese,M.G. (2000) Annotating eukaryote genomes. Curr. Opin. Struct. Biol., 10, 349–354. [DOI] [PubMed] [Google Scholar]
- 4.Eisenberg D., Marcotte,E.M., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. [DOI] [PubMed] [Google Scholar]
- 5.Nakai K. (2000) Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem., 54, 277–344. [DOI] [PubMed] [Google Scholar]
- 6.Bork P., Dandekar,T., Diaz-Lazcoz,Y., Eisenhaber,F., Huynen,M. and Yuan,Y. (1998) Predicting function: from genes to genomes and back. J. Mol. Biol., 283, 707–725. [DOI] [PubMed] [Google Scholar]
- 7.Berman H.M., Westbrook,J., Feng,Z., Gillliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting C.P., Schultz,J., Milpetz,F. and Bork,P. (1999) SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res., 27, 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnhammer E.L., Eddy,S.R. and Durbin,R. (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins, 28, 405–420. [DOI] [PubMed] [Google Scholar]
- 10.Puntervoll P., Linding,R., Gemund,C., Chabanis-Davidson,S., Mattingsdal,M., Hunter,W., Aasland,R. and Gibson,T. (2003) The ELM server: a new resource for revealing short functional sites in modular eukaryotic proteins. Nucleic Acids Res., 31, 3625–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen L.J., Gupta,R., Blom,N., Devos,D., Tamames,J., Kesmir,C., Nielsen,H., Staerfeldt,H.H., Rapacki,K., Workman,C. et al. (2002) Prediction of human protein function from post-translational modifications and localization features. J. Mol. Biol., 319, 1257–1265. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tinland B., Koukolikova-Nicola,Z., Hall,M.N. and Hohn,B. (1992) The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc. Natl Acad. Sci. USA, 89, 7442–7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cokol M., Nair,R. and Rost,B. (2000) Finding nuclear localisation signals. EMBO Reports, 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair R., Carter,P. and Rost,B. (2003) NLSdb: database of nuclear localization signals. Nucleic Acids Res., 31, 397–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nair R. and Rost,B. (2002) Inferring sub-cellular localization through automated lexical analysis. Bioinformatics, 18 (Suppl. 1), S78–S86. [DOI] [PubMed] [Google Scholar]
- 18.Dasarathy B.V. (1991) Nearest Neighbor (NN) Norms: NN Pattern Classification Techniques. <IEEE Computer Society Press, Las Alamitos, CA. [Google Scholar]
- 19.Yang Y. and Liu,X. (1999) A re-examination of text categorisation methods. Proceedings of the ACM SIGIR Conference on Research and Development in Information Retrieval, ACM, New York, NY, pp. 42–49. [Google Scholar]
- 20.Nair R. and Rost,B. (2002) Sequence conserved for subcellular localization. Protein Sci., 11, 2836–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenhaber F. and Bork,P. (1998) Wanted: subcellular localization of proteins based on sequence. Trends Cell Biol., 8, 169–170. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S., Madden,T., Shaffer,A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D. (1997) Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sander C. and Schneider,R. (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 25.Rost B. (1999) Twilight zone of protein sequence alignments. Protein Eng., 12, 85–94. [DOI] [PubMed] [Google Scholar]
- 26.Sander C. and Schneider,R. (1991) Database of homology-derived structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 27.Wall L. and Schwartz,R.L. (1990) Programming Perl. O'Reilly and Associates, Inc., Sebastopol, CA. [Google Scholar]
- 28.Stajich J.E., Block,D., Boulez,K., Brenner,S.E., Chervitz,S.A., Dagdigian,C., Fuellen,G., Gilbert,J.G., Korf,I., Lapp,H. et al. (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res., 12, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein L.D. (2001) Using Perl to facilitate biological analysis. Methods Biochem. Anal., 43, 413–449. [DOI] [PubMed] [Google Scholar]
- 30.Carter P., Liu,J. and Rost,B. (2003) PEP: predictions for entire proteomes. Nucleic Acids Res., 31, 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]