Abstract
NEWT is a new taxonomy portal to the SWISS-PROT protein sequence knowledgebase. It contains taxonomy data, which is updated daily, for the complete set of species represented in SWISS-PROT, as well as those stored at the NCBI. Users can navigate through the taxonomy tree and access corresponding SWISS-PROT protein entries. In addition, a manually curated selection of external links allows access to specific information on selected species. NEWT is available at http://www.ebi.ac.uk/newt/.
NEWT TAXONOMY DATA
Species denomination
Have you ever been bewildered by items on trendy restaurant menus? Then imagine the puzzled look of a customer who has just been offered ‘Bos taurus filet served on a bed of Phaseolus vulgaris, decorated with a slice of Lycopersicon esculentum and accompanied by Solanum tuberosum’. Whilst those names may not actually appear on restaurant menus, they are the scientific names of beef, French bean, tomato and potato, respectively. In order to group the different names that describe the same organisms, several taxonomy databases have been devised.
The New Taxonomy database of the SWISS-PROT group (NEWT http://www.ebi.ac.uk/newt/) integrates taxonomy data specific to the SWISS-PROT knowledgebase (1) with information provided by the NCBI taxonomic database (2).
Species, for which protein sequence data are available, are named according to the SWISS-PROT nomenclature. The latter usually consists of the Latin scientific name, formed according to the binomial system of Linnaeus, that is, the genus followed by the species (e.g. Cannabis sativa). For most species, the scientific name is followed by the English common name (e.g. hemp) and a synonym when available (e.g. marijuana). Following SWISS-PROT conventions, a systematic approach for naming viral and bacterial strains and isolates has been adopted. Furthermore, the SWISS-PROT Organism Species code (OS code) is also given, i.e. the five-letter mnemonic code which appears in the protein entry identifier of the SWISS-PROT database (e.g. CANSA), as well as the full list of synonyms stored in the NCBI database. Users can refer to the corresponding data via a link to the NCBI taxonomy server.
Taxonomic lineage
Taxonomy is organized in a tree structure, which represents the taxonomic lineage. The position of each node on a tree is determined by its rank in the taxonomy hierarchy, so that the last ranks (usually species or sub-species) represent the ‘leaves’ on the tree's branches, and higher ranks like ‘phylum’, ‘order’ and ‘family’ are placed higher on the tree. The ordered list of the nodes forms the lineage (Table 1). The NEWT database stores the taxonomy tree structure, thus making it possible to navigate from one node to another and to access the lineage for each node.
Table 1. Example of a lineage.
| Node of lineage | NCBI rank | Description |
|---|---|---|
| Eukaryota | Superkingdom | With nucleated cells |
| Metazoa | Kingdom | Multi-cellular organism developed from an embryo |
| Arthropoda | Phylum | With jointed legs |
| Tracheata | No rank | Which breathes by tracheae |
| Hexapoda | Superclass | With six legs |
| Insecta | Class | Insect |
| Pterygota | No rank | Winged insect |
| Neoptera | Subclass | Who is able to fold its wings back, flat over its body |
| Endopterygota | Infraclass | Wings develop internally |
| Hymenoptera | Order | Membrane-winged |
| Apocrita | No rank | Waisted appearance |
| Aculeata | Suborder | With prickles or stings |
| Apoidea | Superfamily | |
| Apidae | Family | |
| Apis | Genus | Bee |
NEWT LINKS
Integration with SWISS-PROT
For every taxon stored in NEWT where protein sequence data in SWISS-PROT or TrEMBL (the computer-annotated supplement of SWISS-PROT) is found, the total number of corresponding entries is indicated. A direct link to the ExPASy server (3) also allows the user to retrieve all protein entries relative to a given taxon. The number of entries is also compiled for higher nodes in the taxonomy tree, so that users can retrieve all bacterial sequences in SWISS-PROT or TrEMBL, for example.
Additionally, the taxonomy information in NEWT can be accessed from the NiceProt view of a protein entry by clicking on the links in the taxonomy section (e.g. http://www.expasy.org/cgi-bin/niceprot.pl?P00053).
External information
It cannot be unfair to say that species are seldom only known by their scientific Latin name, and whilst common names like ‘African elephant’ are familiar to most, others such as ‘Chinese water mocassin’ remain obscure to all save the specialist. Fortunately though, with the explosion of information on the web, the number of high-standard web sites entirely devoted to a particular species has multiplied. NEWT makes use of this kind of resource by providing a manually-curated selection of relevant links to pages and images on foreign web sites. Currently, links are available for over 12 000 taxa.
Also where available, a direct link to the corresponding entry on the NCBI taxonomy web server is provided.
Finally, for species whose complete genome has been sequenced and translated, NEWT provides a link to the Proteome pages at EBI (4).
NEWT SERVICES
NEWT can be searched by species name (either in scientific Latin or English), with an option to use wildcards anywhere in the query (Fig. 1). Alternatively, searches can be conducted using the NCBI unique taxonomy identifier (taxID).
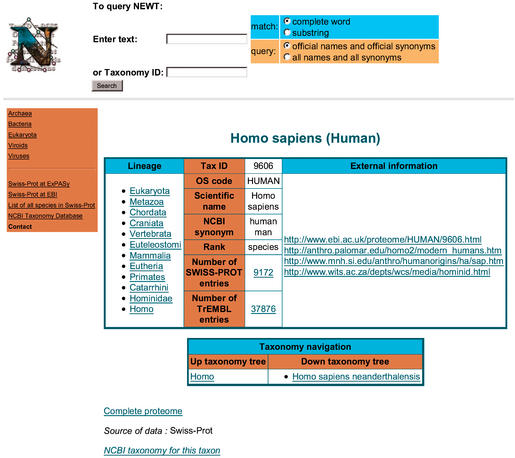
Figure 1.
Entry display on the NEWT web server.
Entries in the results list are flagged when external links are available (Fig. 2). The same flag is used in the navigation table, thus allowing the user to explore the taxonomy tree node by node.
Figure 2.
Search display on the NEWT web server.
Future developments include a ‘species of the day’ link to NEWT from the main ExPASy page.
CONCLUSION
Thanks to the NEWT server, it is now possible to navigate seamlessly between taxonomic information and proteins.
REFERENCES
- 1.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 28, 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gasteiger E., Gattiker,A., Hoogland,C., Ivanyi,I., Appel,R.D. and Bairoch,A. (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res., 31, 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apweiler R., Biswas,M., Fleischmann,W., Kanapin,A., Karavidopoulou,Y., Kersey,P., Kriventseva,E.V., Mittard,V., Mulder,N., Phan,I. and Zdobnov,E. (2001) Proteome Analysis Database: online application of InterPro and CluSTr for the functional classification of proteins in whole genomes. Nucleic Acids Res., 29, 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]