Abstract
PipeAlign is a protein family analysis tool integrating a five step process ranging from the search for sequence homologues in protein and 3D structure databases to the definition of the hierarchical relationships within and between subfamilies. The complete, automatic pipeline takes a single sequence or a set of sequences as input and constructs a high-quality, validated MACS (multiple alignment of complete sequences) in which sequences are clustered into potential functional subgroups. For the more experienced user, the PipeAlign server also provides numerous options to run only a part of the analysis, with the possibility to modify the default parameters of each software module. For example, the user can choose to enter an existing multiple sequence alignment for refinement, validation and subsequent clustering of the sequences. The aim is to provide an interactive workbench for the validation, integration and presentation of a protein family, not only at the sequence level, but also at the structural and functional levels. PipeAlign is available at http://igbmc.u-strasbg.fr/PipeAlign/.
INTRODUCTION
Protein sequence analysis is a key issue in post-genomic biology. High-throughput genome sequencing and assembly techniques, structural proteomics and gene expression analysis have led to a rapid increase in the amount of sequence, structure and functional data available in the public databases. In order to fully understand the biological role of a particular protein, such diverse information as cellular location, 2D/3D structures, mutations and their associated illnesses, the evolutionary context and literature references must be retrieved, validated, classified and made available to the biologist. The integration of the protein in the context of the complete family is an essential first step in the analysis process. As a consequence, a new generation of protein family analysis tools is now required to organise this heterogeneous, often predicted data into a structured, hierarchical network of connected information.
Here, we present the PipeAlign web server, which offers an integrated, interactive approach to protein family analysis. The rationale of the PipeAlign design is the automation of the initial stages of the analysis process, i.e. the retrieval of homologous sequences and other related information and the hierarchical organisation of this information in the context of a multiple alignment of complete sequences (MACS) (1). As the MACS presents a synthetic view of the variability along a sequence and among homologous sequences, it represents an ideal workbench for the integration of the heterogeneous, often predicted data associated with each of the different members of a protein family. In the context of the MACS, the information can be statistically validated, classified and reliably propagated at either the family or the sub-family level, as appropriate.
SOFTWARE MODULES
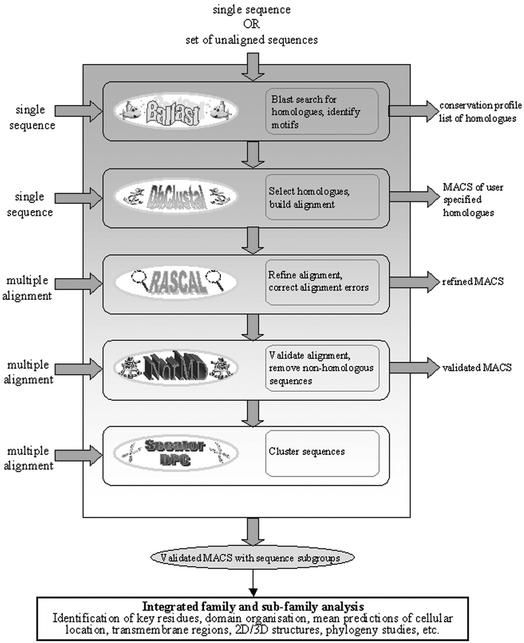
PipeAlign takes either (i) a single protein sequence, (ii) a set of unaligned sequences or (iii) a set of aligned sequences as input and automatically performs a cascade of five different sequence analysis programs, recently developed in-house (Fig. 1). The first task is to perform initial Ballast processing (2) including BLAST database searches (3) and subsequent delineation of the local maximum conserved segments (LMSs). A high-quality MACS of potential homologues is then constructed using the DbClustal multiple alignment program (4) and the RASCAL alignment analysis and correction program (5). Quality validation of the MACS and removal of any sequences that do not belong to the protein family are performed by the NorMD objective function (6). Finally, the sequences are clustered into potential functional subfamilies using two different, complementary programs. By default, the Secator program (7) is used and the DPC program (8) is offered as an optional alternative. Each of the five core programs in PipeAlign is an independent software module and the web interface includes separate entry and exit points at each step in the pipeline.
Figure 1.
The PipeAlign protein family analysis process.
USER INTERFACE
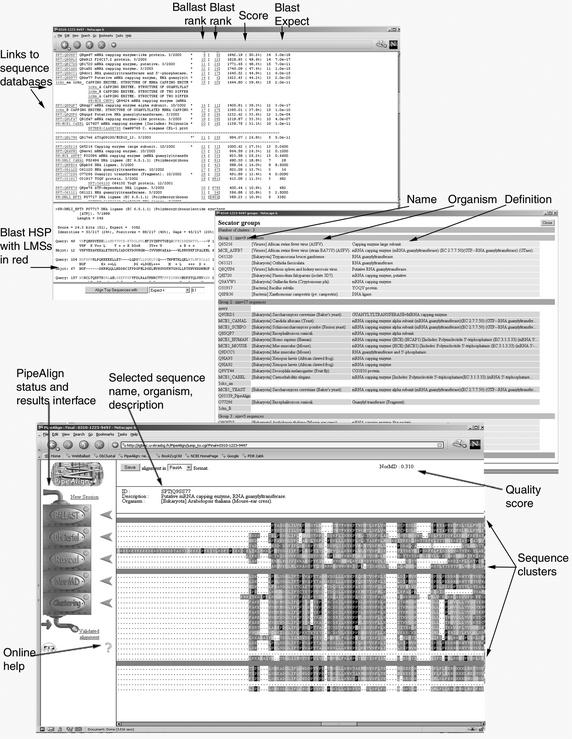
The PipeAlign web server is designed to work in two basic modes (Fig. 1). In the fully automatic mode, a user enters a single sequence or set of unaligned sequences and the complete pipeline of five software modules is launched. When more than one sequence is input, options are provided to align exclusively the user's sequences or to include any additional homologues detected by a database search. In either case, the final result is a high quality MACS of the protein family, which can be viewed with the interactive, graphical browser (Fig. 2). The sequences in the MACS are colour coded according to distinct properties in order to highlight conserved residues and individual family members are clustered into subgroups. Links are provided to relevant information mined during the PipeAlign process, e.g. the local pairwise alignments produced by BLAST and the LMSs deduced by Ballast, the full sequence information in the SWISS-PROT/SpTrEMBL (9) databases as well as 3D structural information in the PDB database (10) when available. Figure 2 shows some of the results available for a typical automatic protein sequence analysis with PipeAlign. In this case, a yeast mRNA guanylyltransferase protein (SWISS-PROT identifier MCE1_YEAST) was used as the query sequence. Thirty-four sequences were automatically selected from the BLAST database searches for alignment with DbClustal, of which three DNA ligase sequences were considered to be unrelated to the query and were subsequently removed from the alignment. The remaining 31 sequences were then clustered into three subgroups: the first mainly composed of kinetoplastidae, bacteria and viruses; the second consisting of metazoa and fungi; and a third subgroup of plants. This automatic process is useful for initial analyses of proteins of unknown function and is particularly suitable for high-throughput, automatic systems, such as genome annotations. However, the PipeAlign web server also provides a more flexible approach, in which the user can choose to enter the pipeline at any one of the five different stages in the PipeAlign process. For example, by starting the PipeAlign process at the DbClustal entry point, it is possible to review the results of the database search and to manually select the set of homologues to be included in the MACS. In addition, PipeAlign provides a number of options for the refinement, validation and clustering of existing multiple sequence alignments, either from other automatic methods or manually edited.
Figure 2.
Screenshots of a typical PipeAlign automa.
The complexity of the PipeAlign analysis process means that the default parameters used at each stage may not necessarily be the most suitable parameters for the particular protein family studied. The choice of query sequence for the initial database search, as well as the threshold used to select proteins for inclusion in the final multiple alignment are crucial to the success of the PipeAlign analysis. The web server therefore offers the biologist the possibility to review the results of each step in the process, to modify certain key parameters and, if necessary, to launch the subsequent software modules. This allows an evaluation and eventual correction of the PipeAlign results and facilitates the analysis and integration of the consequent functional, structural and evolutionary inferences. Such detailed analyses based on MACS can yield important new structural and/or functional insights and form the basis for new hypotheses that can then be tested experimentally. The accuracy and reliability of the PipeAlign analysis has recently been exploited in a number of different projects, from the comparison of three complete genomes of hyperthermophilic archaea (11) and the semi-automatic annotation of the Pyrococcus abyssi genome (12) to the in-depth study of ribosomal genes in 66 different complete genomes (13).
QUALITY CONTROL
An important aspect of the PipeAlign design is the incorporation of quality control procedures at each stage in the analysis process. The initial processing of the BLAST database search results by Ballast uses only those sequences with high significance scores (E<0.1) for the construction of the conservation profile, used in the detection of the LMSs. The LMSs represent locally conserved segments which can be used as reliable anchor points for the DbClustal multiple alignment program. The result is a high-quality global multiple alignment of the full-length sequences, even for highly divergent sequences with large N/C-terminal extensions or internal insertions. Nevertheless, local misalignments can still occur and for this reason, the RASCAL (rapid scanning and correction of alignments) program is used to detect potential badly aligned regions and refine them. The final validation of the MACS alignment is performed by the NorMD objective function. Any sequences in the MACS which are considered to be unrelated to the initial query sequence are removed at this stage.
PERSPECTIVES
While the parameters used in the PipeAlign modules have been selected to be suitable for the majority of protein families, further studies are required to determine the optimal parameters for special cases, such as proteins with a bias in their residue composition. Future developments will also include a hierarchical conservation analysis package and integration with a 3D display. A data mining module is also being developed to provide access to external databases such as the Interpro database (14). This will allow the user to visualise a selection of properties, including known protein domains, motifs and secondary structures.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpital Universitaire de Strasbourg and the Fond National de la Science (GENOPOLE).
REFERENCES
- 1.Lecompte O., Thompson,J.D., Plewniak,F., Thierry,J. and Poch,O. (2001) Multiple alignment of complete sequences (MACS) in the post-genomic era. Gene, 270, 17–30. [DOI] [PubMed] [Google Scholar]
- 2.Plewniak F., Thompson,J.D. and Poch,O. (2000) Ballast: blast post-processing based on locally conserved segments. Bioinformatics, 9, 750–759. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 17, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson J.D., Plewniak,F., Thierry,J. and Poch,O. (2000) DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res., 15, 2919–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson J.D., Plewniak,F., Thierry,J. and Poch,O. (2003) RASCAL: Rapid scanning and correction of multiple sequence alignment programs. Bioinformatics, in press. [DOI] [PubMed] [Google Scholar]
- 6.Thompson J.D., Plewniak,F., Ripp,R., Thierry,J.C. and Poch,O. (2001) Towards a reliable objective function for multiple sequence alignments. J. Mol. Biol., 4, 937–951. [DOI] [PubMed] [Google Scholar]
- 7.Wicker N., Perrin,G.R., Thierry,J.C. and Poch,O. (2001) Secator: a program for inferring protein subfamilies from phylogenetic trees. Mol. Biol. Evol., 8, 1435–1441. [DOI] [PubMed] [Google Scholar]
- 8.Wicker N., Dembele,D., Raffelsberger,W. and Poch,O. (2002) Density of points clustering, application to transcriptomic data analysis. Nucleic Acids Res., 18, 3992–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 1, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sussman J.L., Lin,D., Jiang,J., Manning,N.O., Prilusky,J., Ritter,O. and Abola,E.E. (1998) Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr., 54, 1078–1084. [DOI] [PubMed] [Google Scholar]
- 11.Lecompte O., Ripp,R., Puzos-Barbe,V., Duprat,S., Heilig,R., Dietrich,J., Thierry,J.C. and Poch,O. (2001) Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic archaea. Genome Res., 6, 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen G., Barbe,V., Flament,D., Galperin,M., Heilig,R., Lecompte,O., Poch,O., Prieur,D., Quérellou,J., Ripp,R., et al. (2003) An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol., 47, 1495–1512. [DOI] [PubMed] [Google Scholar]
- 13.Lecompte O., Ripp,R., Thierry,J.C., Moras,D. and Poch,O. (2002) Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res., 24, 5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P., Bucher,P., et al. (2002) InterPro: an integrated documentation resource for protein families, domains and functional sites. Brief Bioinform., 3, 225–235. [DOI] [PubMed] [Google Scholar]