Abstract
SWISS-MODEL (http://swissmodel.expasy.org) is a server for automated comparative modeling of three-dimensional (3D) protein structures. It pioneered the field of automated modeling starting in 1993 and is the most widely-used free web-based automated modeling facility today. In 2002 the server computed 120 000 user requests for 3D protein models. SWISS-MODEL provides several levels of user interaction through its World Wide Web interface: in the ‘first approach mode’ only an amino acid sequence of a protein is submitted to build a 3D model. Template selection, alignment and model building are done completely automated by the server. In the ‘alignment mode’, the modeling process is based on a user-defined target-template alignment. Complex modeling tasks can be handled with the ‘project mode’ using DeepView (Swiss-PdbViewer), an integrated sequence-to-structure workbench. All models are sent back via email with a detailed modeling report. WhatCheck analyses and ANOLEA evaluations are provided optionally. The reliability of SWISS-MODEL is continuously evaluated in the EVA-CM project. The SWISS-MODEL server is under constant development to improve the successful implementation of expert knowledge into an easy-to-use server.
INTRODUCTION
Three-dimensional (3D) protein structures provide valuable insights into the molecular basis of protein function, allowing an effective design of experiments, such as site-directed mutagenesis, studies of disease-related mutations or the structure based design of specific inhibitors. Although great progress was made in the field of experimental structure solution by X-ray crystallography and nuclear magnetic resonance spectroscopy (NMR), it is still a time-consuming process without guaranteed success. Currently, about 20 000 experimental protein structures are deposited in the Protein Data Bank (PDB) (1). Nevertheless, the number of structurally characterized proteins is low compared to the number of known protein sequences: taken together, the SWISS-PROT and TrEMBL databases hold about 850 000 sequence entries (2), which exceeds the number of known different structures by about two orders of magnitude. Thus, no structural information is available for the vast majority of protein sequences. Therefore, it is an obvious demand to bridge this ‘structure knowledge gap’ and computational methods for protein structure prediction have gained much interest in recent years.
Among all current theoretical approaches, comparative modeling is the only method that can reliably generate a 3D model of a protein (target) from its amino acid sequence (3,4). Successful model building requires at least one experimentally solved 3D structure (template) that has a significant amino acid sequence similarity to the target sequence. Various structural genomics initiatives were started in the last few years, aiming to speed up the elucidation of new protein structures (5). Experimental structure elucidation and comparative modeling complement one another in the exploration of the protein structure space. A key to the efficient coverage will be the careful and optimal selection of the proteins for structural genomics (6). The growing number of structural templates brings a steadily increasing number of sequences into ‘modeling distance’ for comparative modeling.
Modeling of protein structures usually requires extensive expertise in structural biology and the use of highly specialized computer programs for each of the individual steps of the modeling process (3). The idea of an easy-to-use, automated modeling facility with integrated expert knowledge was first implemented 10 years ago by Peitsch et al. (7–9) and formed the starting point for the SWISS-MODEL server. Since then, SWISS-MODEL has been continuously developed, and is today the most widely used modeling server publicly available on the web (10–12). In 2002 the server computed 120 000 user requests for 3D protein models. In recent years, several other groups have developed similar systems for automated homology modeling [e.g. ModPipe (http://www.salilab.org) (13), CPHmodels (http://www.cbs.dtu.dk/services/CPHmodels/) (14), 3D-JIGSAW (http://www.bmm.icnet.uk/~3djigsaw/) (15), ESyPred3D (http://www.fundp.ac.be/urbm/bioinfo/esypred/) (16) or SDSC1 (http://cl.sdsc.edu/hm.html)].
SWISS-MODEL MODES
The SWISS-MODEL server is designed to work with a minimum of user input, i.e. in the simplest case, only the amino acid sequence of a target protein. As comparative modeling projects can be of different complexity, additional user input may be necessary for some modeling projects, e.g. to select a different template or adjust the target-template alignment. Therefore, the SWISS-MODEL server gives the user the choice between three main interaction modes.
First approach mode
The ‘first approach mode’ provides a simple interface and requires only an amino acid sequence as input data. The server will automatically select suitable templates. Optionally, the user can specify up to five template structures, either from the ExPDB library or uploaded coordinate files. The automated modeling procedure will start if at least one modeling template is available that has a sequence identity of more than 25% with the submitted target sequence. However, users need to be aware that the model reliability decreases as the sequence identity decreases and that target-template pairs sharing less than 50% sequence identity may often require manual adjustment of the alignment (see MODELING RESULTS AND EVALUATION).
Alignment mode
In the ‘alignment mode’ the modeling procedure is initiated by submitting a sequence alignment. The user specifies which sequence in the given alignment is the target sequence and which one corresponds to a structurally known protein chain from the ExPDB template library. The server will build the model based on the given alignment.
Project mode
The ‘project mode’ allows the user to submit a manually optimized modeling request to the SWISS-MODEL server. The starting point for this mode is a DeepView project file. It contains the superposed template structures, and the alignment between the target and the templates. This mode gives the user control over a wide range of parameters, e.g. template selection or gap placement in the alignment. Furthermore, the project mode can also be used to iteratively improve the output of the ‘first approach mode’.
DeepView—Swiss-PdbViewer
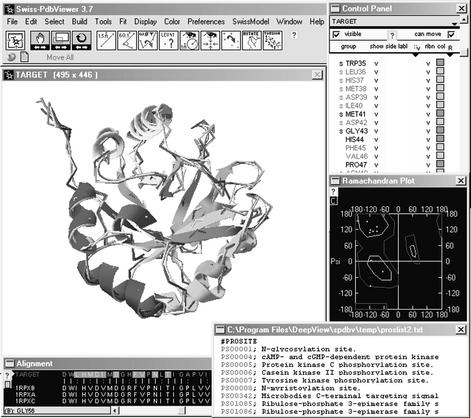
The program DeepView (Swiss-PdbViewer) was designed to integrate functions for protein structure visualization, analysis and manipulation into a sequence-to-structure workbench with a user-friendly interface. It allows the user to manage complex modeling projects and is publicly available from the ExPASy server (http://www.expasy.org/spdbv). With DeepView one can search for suitable modeling templates and download the corresponding PDB or ExPDB files directly from the DeepView server. Using the integrated sequence alignment tools and structural superposition algorithms, a target sequence can be mapped onto the modeling templates in one step. Then the initial sequence alignment can be optimized manually while the anticipated changes in the model backbone are reflected in real-time in the displayed structural superposition. The complete project file is then submitted to the SWISS-MODEL server for model building. The resulting protein model can be visualized and analyzed using the integrated tools (Fig. 1).
Figure 1.
SWISS-MODEL project displayed in DeepView. Project files contain the final model coordinates, accompanied by the superposed template structures. Adjustments of the underlying target-template alignment can be done manually. DeepView provides several tools to visualize and analyze the modeling results.
MODELING PROCEDURE
All homology-modeling methods consist of the following four steps: (i) template selection; (ii) target template alignment; (iii) model building; and (iv) evaluation. These steps can be iteratively repeated, until a satisfying model structure is achieved. Several different techniques for model building have been developed (11,14,17,18). The SWISS-MODEL server approach can be described as rigid fragment assembly [first implemented in Composer (18)], which will be outlined briefly.
Template selection
The SWISS-MODEL server template library ExPDB is extracted from the PDB (1). In order to allow a stable and automated workflow of the server, the PDB coordinate files are split into individual protein chains and unreliable entries, e.g. theoretical models and low quality structures providing only Cα coordinates, are removed. Additional information useful for template selection is gathered and added to the file header, e.g. probable quaternary structure (19), quality indicators like empirical force field energy (20) or ANOLEA mean force potential scores (21). To select templates for a given protein, the sequences of the template structure library are searched (22,23). If these templates cover distinct regions of the target sequence, the modeling process will be split into separate independent batches.
Alignment
Up to five template structures per batch are superposed using an iterative least squares algorithm. A structural alignment is generated after removing incompatible templates, i.e. omitting structures with high Cα root mean square deviations to the first template. A local pair-wise alignment of the target sequence to the main template structures is calculated (24), followed by a heuristic step to improve the alignment for modeling purposes. The placement of insertions and deletions is optimized considering the template structure context. In particular, isolated residues in the alignment (‘islands’) are moved to the flanks to facilitate the loop building process.
Model building
To generate the core of the model, the backbone atom positions of the template structure are averaged. The templates are thereby weighted by their sequence similarity to the target sequence, while significantly deviating atom positions are excluded. The template coordinates cannot be used to model regions of insertions or deletions in the target-template alignment. To generate those parts, an ensemble of fragments compatible with the neighboring stems is constructed using constraint space programming (CSP). The best loop is selected using a scoring scheme, which accounts for force field energy, steric hindrance and favorable interactions like hydrogen bond formation. If no suitable loop can be identified, the flanking residues are included to the rebuilt fragment to allow for more flexibility. In cases where CSP does not give a satisfying solution and for loops above 10 residues, a loop library derived from experimental structures is searched to find compatible loop fragments.
Side chain modeling
The reconstruction of the model side chains is based on the weighted positions of corresponding residues in the template structures. Starting with conserved residues, the model side chains are built by iso-sterically replacing template structure side chains. Possible side chain conformations are selected from a backbone dependent rotamer library, which has been constructed carefully taking into account the quality of the source structures (25). A scoring function assessing favorable interactions (hydrogen bonds, disulfide bridges) and unfavorably close contacts is applied to select the most likely conformation.
Energy minimization
Deviations in the protein structure geometry, which have been introduced by the modeling algorithm when joining rigid fragments are regularized in the last modeling step by steepest descent energy minimization using the GROMOS96 force field (20). Empirical force fields are useful to detect parts of the model with conformational errors. In our own experience and the work of others (26,27), energy minimization or molecular dynamics methods are in general not able to improve the accuracy of the models, and are used in SWISS-MODEL only to regularize the structure. However, the successful application of restricted molecular dynamics for improving homology models has recently been reported for a few test cases (28). To derive more general rules of engagement of molecular dynamics, further systematic experiments have to be conducted.
The four modeling steps—template superposition, target-template alignment, model building and energy minimization—have been implemented in the program ProModII in ANSI C (11).
MODELING RESULTS AND EVALUATION
Possible applications of protein models depend largely on the quality of the models (29–31). The accuracy of a model can vary significantly, even within different regions of the same protein: usually highly-conserved core regions can be modeled much more reliably than variable loop regions or surface residues. Several tools are provided to allow the SWISS-MODEL user to evaluate the reliability of the model: similar to B-factors in crystal structures, the corresponding column in SWISS-MODEL result files consists of a C-score, which gives an estimate of the variability of the template structures at this position. Parts of the model where no template information could be used for model building (insertions or deletions) are assigned a C-score of 99. A detailed log file listing all steps performed by the modeling server is provided to the user. This includes force field energy (20) for the overall structure and for each individual residue to identify regions with obvious conformational or electrostatic problems. Optionally, WhatCheck (32) reports and evaluation by the atomic mean force potential ANOLEA (21) are provided by SWISS-MODEL to assess the quality of the model.
Inaccurate target-template alignments are the most frequent source of errors in models. This is especially true when the sequence similarity between the target and the template sequence drops below 40%, and manual editing of the alignment is necessary to achieve a satisfying model. The program DeepView (Swiss-PdbViewer) (10,11) can be used to revise the resulting SWISS-MODEL projects from ‘first approach’ mode submissions. DeepView allows users to manually adjust the alignment while visually verifying the structural implications, e.g. the placement of insertions and deletions in the correct structural context or the conservation of structural features with a functional role. The modified modeling project is then resubmitted for another round of model building to the server via the ‘project mode’. Finally, fine-tuning of the model, such as energy checks, loop building and rotamer search can also be performed directly on the returned project files with DeepView.
Server evaluation
During the first large-scale modeling experiment named 3D-Crunch in 1998 (10), in which over 200 000 protein sequences were submitted to the SWISS-MODEL pipeline, a control set of 1200 sequences with known structures was used to assess the reliability of SWISS-MODEL. For instance, 79% of the sequences sharing 50–59% identity with their templates yielded models with Cα positions deviating by less than 3 Å compared to their experimental crystal structures (12). These results were used to optimize the procedures for the fully automatic mode of the server. The continuous automated blind evaluation of servers such as SWISS-MODEL by EVA-CM (33) provides detailed reports about the accuracy and reliability of automated modeling servers. Results are available publicly on the web and allow users to estimate the expected accuracy of different modeling methods.
ACCESS TO SERVER AND SOFTWARE
SWISS-MODEL is accessible via a web interface at http://swissmodel.expasy.org, or directly as a link from SWISS-PROT (2) entries on the ExPASy server (34). The program DeepView (Swiss-PdbViewer) can be downloaded for free at http://www.expasy.org/spdbv/. Depending on the complexity of the modeling task and server workload, it may take a few minutes to several hours for the server to build a model, including energy minimization. The model coordinates and log-files are returned to the user by email. The computational resources for the SWISS-MODEL server are provided by a collaboration between the Swiss Institute of Bioinformatics at the Biozentrum Basel (University of Basel, Switzerland) and the Advanced Biomedical Computing Center (NCIFCRF Frederick, MD, USA).
OUTLOOK
It is expected that protein crystallography and NMR will provide 3D structures for at least one representative for 90% of all protein domains within the next decade (6). This will enable comparative modeling to build protein models for the vast majority of sequences. Additional benefit will be achieved by combining sequence-based functional information with protein 3D information. In an ongoing project, 3D models built by SWISS-MODEL are integrated into the INTERPRO (35) database. Making protein models easily available is one of the great advantages of automated comparative protein modeling. As a consequence, today's sequence centered view on proteins will continuously evolve to a more complete picture including protein structure information. SWISS-MODEL will be continuously developed to improve the successful implementation of expert knowledge into an easy-to-use homology-modeling server.
Acknowledgments
ACKNOWLEDGEMENTS
We are indebted to Elisabeth Gasteiger (SIB Geneva, Switzerland), Karol Miaskiewicz and Jack Collins (NCIFCRF Frederick, MD, USA) for excellent technical support. We are especially grateful to Nicola Mulder and Rolf Apweiler (EBI Hinxton, UK) for encouraging discussions. We thank Alexander Diemand (SIB Lausanne, Switzerland) for his contributions to the Linux version of DeepView.
REFERENCES
- 1.Westbrook J., Feng,Z., Chen,L., Yang,H. and Berman,H.M. (2003) The Protein Data Bank and structural genomics. Nucleic Acids Res., 31, 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.-C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tramontano A., Leplae,R. and Morea,V. (2001) Analysis and assessment of comparative modeling predictions in CASP4. Proteins, 45 (Suppl. 5), 22–38. [DOI] [PubMed] [Google Scholar]
- 4.Marti-Renom M.A., Stuart,A.C., Fiser,A., Sanchez,R., Melo,F. and Sali,A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct., 29, 291–325. [DOI] [PubMed] [Google Scholar]
- 5.Brenner S.E. (2001) A tour of structural genomics. Nature Rev. Genet., 2, 801–809. [DOI] [PubMed] [Google Scholar]
- 6.Vitkup D., Melamud,E., Moult,J. and Sander,C. (2001) Completeness in structural genomics. Nature Struct. Biol., 8, 559–566. [DOI] [PubMed] [Google Scholar]
- 7.Peitsch M.C. and Jongeneel,C.V. (1993) A 3-D model for the CD40 ligand predicts that it is a compact trimer similar to the tumor necrosis factors. Intern. Immunol., 5, 233–238. [DOI] [PubMed] [Google Scholar]
- 8.Peitsch M.C. (1995) Protein modelling by E-Mail. BioTechnology, 13, 658–660. [Google Scholar]
- 9.Peitsch M.C. (1996) ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans., 24, 274–279. [DOI] [PubMed] [Google Scholar]
- 10.Guex N., Diemand,A. and Peitsch,M.C. (1999) Protein modelling for all. Trends Biochem. Sci., 24, 364–367. [DOI] [PubMed] [Google Scholar]
- 11.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 12.Schwede T., Diemand,A., Guex,N. and Peitsch,M.C. (2000) Protein structure computing in the genomic era. Res. Microbiol., 151, 107–112. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez R. and Sali,A. (1998) Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl Acad. Sci. USA, 95, 13597–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lund O., Frimand,K., Gorodkin,J., Bohr,H., Bohr,J., Hansen,J. and Brunak,S. (1997) Protein distance constraints predicted by neural networks and probability density functions. Protein Eng., 10, 1241–1248. [DOI] [PubMed] [Google Scholar]
- 15.Bates P.A., Kelley,L.A., MacCallum,R.M. and Sternberg,M.J. (2001) Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins, 45 (Suppl. 5), 39–46. [DOI] [PubMed] [Google Scholar]
- 16.Lambert C., Leonard,N., De Bolle,X. and Depiereux,E. (2002) ESyPred3D: Prediction of proteins 3D structures. Bioinformatics, 18, 1250–1256. [DOI] [PubMed] [Google Scholar]
- 17.Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 18.Sutcliffe M.J., Haneef,I., Carney,D. and Blundell,T.L. (1987) Knowledge based modelling of homologous proteins, Part I: Three-dimensional frameworks derived from the simultaneous superposition of multiple structures. Protein Eng., 1, 377–384. [DOI] [PubMed] [Google Scholar]
- 19.Henrick K. and Thornton,J.M. (1998) PQS: a protein quaternary structure file server. Trends Biochem. Sci., 23, 358–361. [DOI] [PubMed] [Google Scholar]
- 20.van Gunsteren W.F., Billeter,S.R., Eising,A., Hünenberger,P.H., Krüger,P., Mark,A.E., Scott,W.R.P. and Tironi,I.G. (1996) Biomolecular Simulations: The GROMOS96 Manual and User Guide. VdF Hochschulverlag ETHZ, Zürich. [Google Scholar]
- 21.Melo F. and Feytmans,E. (1998) Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol., 277, 1141–1152. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X. and Miller,W. (1991) A time-efficient, linear-space local similarity algorithm. Adv. Appl. Math., 12, 337–357. [Google Scholar]
- 25.Lovell S.C., Word,J.M., Richardson,J.S. and Richardson,D.C. (2000) The penultimate rotamer library. Proteins, 40, 389–408. [PubMed] [Google Scholar]
- 26.Koehl P. and Levitt,M. (1999) A brighter future for protein structure prediction. Nature Struct. Biol., 6, 108–111. [DOI] [PubMed] [Google Scholar]
- 27.Moult J. (1996) The current state of the art in protein structure prediction. Curr. Opin. Biotechnol., 7, 422–427. [DOI] [PubMed] [Google Scholar]
- 28.Flohil J.A., Vriend,G. and Berendsen,H.J. (2002) Completion and refinement of 3-D homology models with restricted molecular dynamics: application to targets 47, 58, and 111 in the CASP modeling competition and posterior analysis. Proteins, 48, 593–604. [DOI] [PubMed] [Google Scholar]
- 29.Sali A. and Kuriyan,J. (1999) Challenges at the frontiers of structural biology. Trends Cell Biol., 9, M20–M24. [PubMed] [Google Scholar]
- 30.Peitsch M.C. (2002) About the use of protein models. Bioinformatics, 18, 934–938. [DOI] [PubMed] [Google Scholar]
- 31.Kopp J., Peitsch,M.C. and Schwede,T. (2003) Protein structure modeling. In Galperin,M.Y. and Koonin,E.V. (eds), Frontiers in Computational Genomics. Caister Academic Press, Norfolk, Vol. 3, pp. 89–121. [Google Scholar]
- 32.Hooft R.W., Vriend,G., Sander,C. and Abola,E.E. (1996) Errors in protein structures. Nature, 381, 272. [DOI] [PubMed] [Google Scholar]
- 33.Eyrich V.A., Marti-Renom,M.A., Przybylski,D., Madhusudhan,M.S., Fiser,A., Pazos,F., Valencia,A., Sali,A. and Rost,B. (2001) EVA: continuous automatic evaluation of protein structure prediction servers. Bioinformatics, 17, 1242–1243. [DOI] [PubMed] [Google Scholar]
- 34.Appel R.D., Bairoch,A. and Hochstrasser,D.F. (1994) A new generation of information retrieval tools for biologists: the example of the ExPASy WWW server. Trends Biochem. Sci., 19, 258–260. [DOI] [PubMed] [Google Scholar]
- 35.Mulder N.J., Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]