Abstract
RNA molecules play an important role in many biological activities. Knowing its secondary structure can help us better understand the molecule's ability to function. The methods for RNA structure determination have traditionally been implemented through biochemical, biophysical and phylogenetic analyses. As the advance of computer technology, an increasing number of computational approaches have recently been developed. They have different goals and apply various algorithms. For example, some focus on secondary structure prediction for a single sequence; some aim at finding a global alignment of multiple sequences. Some predict the structure based on free energy minimization; some make comparative sequence analyses to determine the structure. In this paper, we describe how to correctly use GPRM, a genetic programming approach to finding common secondary structure elements in a set of unaligned coregulated or homologous RNA sequences. GPRM can be accessed at http://bioinfo.cis.nctu.edu.tw/service/gprm/.
INTRODUCTION
It is known that RNAs perform a wide variety of biological activities, ranging from enzyme-like catalysis to protein synthesis. Many of the tasks are accomplished by the binding of proteins to specific sites in RNA molecules (1–3). However, unlike DNA binding proteins, which recognize binding sites of conserved sequences, RNA protein binding sites are more conserved in structures than in sequences. Knowing RNA structures can help us not only gain a deeper insight of the regulation activities, but also identify new members of a specific coregulated RNA family. There have been many various computational methods developed for RNA secondary structure prediction. According to the number of RNA sequences for which to predict the secondary structure, a method can be considered as single-sequence or multiple-sequence structure prediction. The goal of single-sequence structure prediction is to find the possible folding of a single RNA sequence (4–8), while the goal of multiple-sequence structure prediction falls into two categories. One category is focused on finding a global structure alignment (9,10); the other concentrates on common structure element prediction (11–14). In this paper, we describe GPRM, a tool specifically designed to identify common secondary structure elements within a set of homologous or functionally related RNA sequences.
GPRM operates on a population of possible RNA structure elements. With the wealth of structure information kept in a population and the flexibility of genetic operators, GPRM utilizes genetic programming to explore the search space of RNA secondary structure elements. It has been tested on some datasets previously used to verify other prediction systems, and on pseudoknots that most current systems cannot deal with (14). In the following sections, we describe its purpose and limitations, the sequence input, the parameters and finally the output.
MATERIALS AND METHODS
Purpose and limitations
Unlike most current approaches, we consider structure prediction as a supervised learning problem. Given pre-classified training examples, supervised learning is to learn a discriminative concept that distinguishes the examples of different classes. GPRM treats the given family of coregulated RNA sequences as positive examples and negative examples are the sequences randomly generated based on the observed frequencies of sequence alphabet in positive examples. GPRM is aimed at the structure elements that can be used to distinguish the given family from random sequences. As a sufficiently large family is required to better justify the structure elements learned, it is not appropriate to apply GPRM to a single RNA sequence or a small dataset, e.g. a family of fewer than 10 sequences. The elements found by GPRM from a single sequence or a small dataset may be meaningless.
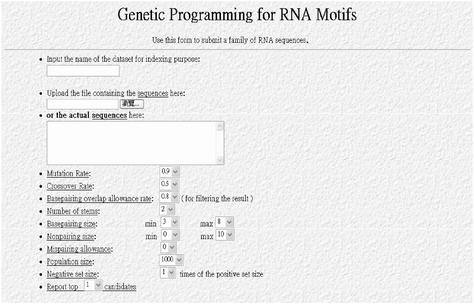
GPRM is a stochastic optimization process. It involves an attempt to optimize a fitness function by modifying and combining tentative structure elements in a population. Due to the non-deterministic characteristics, it is difficult to estimate GPRM's running time in advance. The required CPU time may vary from seconds to hours or even days, depending on the complexity of the target structure elements. The advantage of GPRM lies in the fact that in its entire process, there is no difference between the handling of simple stem-loop structures and any other more complex structures such as pseudoknots. Though it does not guarantee the optimal solution or the same result from multiple runs, GPRM gives the approximately best answers. Its home page is shown in Figure 1.
Figure 1.
The webpage of GPRM. Users can click on the parameters, e.g. Mutation Rate, to see a brief explanation.
Sequence input and parameters
For the time being, GPRM only accepts RNA sequences in the FASTA format. Any blank (‘ ’) or dash (‘–’) in a sequence is ignored, and symbols other than A, G, C, T/U are reported as errors. The length of each sequence is limited to 1000 nt and the total number of nucleotides in a given set cannot exceed 60 000, owing to the hardware limitation of our current PC server. All the constraints can be relaxed after we transfer GPRM to other high-end computer platforms. The input sequences can be either uploaded from the user's local disks or directly typed into a small window on the web page. For the purpose of reference, an additional name should be assigned to the sequence dataset.
Users are required to select the value of several parameters before running GPRM. These parameters are used to either specify the configuration of a structure element or control the evolutionary process in GPRM. All the parameters are detailed as follows.
Basepairning size.
This parameter is used to specify the range of stem lengths. A wider range can represent more candidate elements, but it also increases the running time. On the other hand, a narrow range may over-constrain the search space and consequently make GPRM converge prematurely to wrong structures. The background knowledge of structure elements helps us specify an appropriate range. However, the correct stem length is often unknown beforehand. In case of no background knowledge, we suggest users run GPRM multiple times using various ranges. An appropriate range can usually be inferred from multiple results.
Nonpairing size.
Nonpairing size specifies the range of loop lengths. Similar to basepairing, the correct nonpairing size is generally unknown. Therefore, when background knowledge is limited, we advise users to run GPRM with different loop lengths and later derive the appropriate size from the results.
Number of stems.
This parameter specifies the number of stems in the target structure. Like the two parameters above, the correct parameter value is usually unknown in advance. Users can run GPRM with different numbers when no background knowledge is available. The exact number of stems in the target structure element can be obtained from the comparison of the results.
Mismatch allowance.
In addition to canonical A–U and G–C base pairs, mispaired bases also occur in RNA secondary structures, e.g. the G–U pair. This parameter specifies the maximal number of mispaired bases in a stem other than the G–U pair. The default is zero, which means each base pair in a stem must be either a canonical pair or the G–U pair. If the allowance is set to two, at most two mismatches (other than G–U) are allowed in each stem of the structure element.
Basepairing overlap allowance.
A structure element may occur multiple times in a sequence. Two occurrences are considered overlapped if any stem of one occurrence is overlapped with a stem of the other. The basepairing overlap allowance limits the size of the overlapping part. The lower allowance, the smaller overlapping part permitted. GPRM uses this parameter to filter out spurious structure element occurrences. When the overlapping part exceeds the allowance, the occurrence with shorter stems is discarded.
Negative set size.
Based on supervised learning, GPRM tries to identify in a given RNA family significant consensus structure elements that can be used to distinguish the family members from non-members (15). Random sequences are treated as non-members that form the negative set, and the members of a given family form the positive set. The negative set can be one to five times as large as the given family. If the number of family members is relatively small (e.g. <15), it is recommended to use a larger negative set. A larger negative set can represent non-members more realistically, but it also increases GPRM's running time.
Top candidates.
This parameter specifies how many candidate structure elements are reported. These candidates are ranked by the fitness value. The definition of the fitness function is detailed in (14). Note that since the fitness value does not always reflect the true biological significance, the top-ranked candidate may not be the correct answer. Therefore, we suggest users examine more than one candidate.
Mutation rate.
GPRM's mutation operator changes the configuration of a structure element selected from the population to simulate mutation in nature that causes sporadic and random alterations in genetic materials. Mutation is a non-deterministic process. We use the mutation rate to represent its probability. In GPRM, mutation is the primary operator to optimize potential structure elements by modifying their configurations. The default mutation rate is set as high as 0.9 to encourage more mutation operations.
Crossover rate.
Unlike mutation, GPRM's crossover operation is performed on two elements randomly picked from the population. Its purpose is to exchange the configuration between two tentative elements to generate two better offspring. The crossover rate is the probability that GPRM performs crossover operation. Since the current version of GPRM only allows configuration exchange between two elements with similar structures, the default crossover rate is set at 0.5 only.
Population size.
To constrain the search space, GPRM only operates on a population of tentative structure elements. However, when the search space of candidate elements increases, e.g. owing to a larger range of the basepairing size, the population size needs to be increased as well for accommodating more various elements to avoid overlooking crucial candidates.
Output
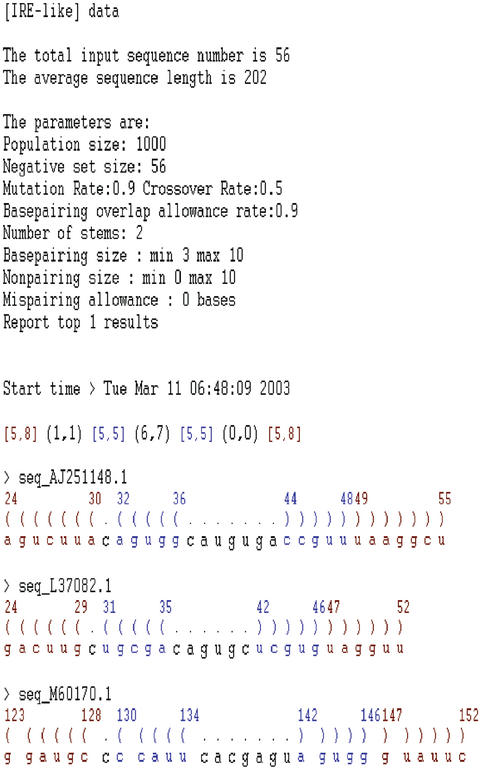
A sample partial output of GPRM is presented in Figure 2. There are three parts in an output. The first part includes the dataset name (e.g. ‘test IRE’ in Fig. 2) for reference and the parameter values specified by the user. The second part is the predicted common structure elements in the given family. GPRM represents a structure element with two kinds of segments, pairing or nonpairing. It uses brackets and parentheses to indicate a pairing segment and a nonpairing segment, respectively. The numbers in brackets and parentheses present the range of segment lengths, e.g. [5,8] means the length of the pairing segment (i.e. a stem) is between five and eight nucleotides. The pairing relation is simply illustrated by color, e.g. two ‘[5,8]’ segments in red are paired as shown in Figure 2. The remaining output is the list of the element occurrences within each sequence in the conventional paired parenthesis format. It shows the starting and the ending position of each pairing segment as well as the subsequence that form the secondary structure.
Figure 2.
A sample output of GPRM. An output is divided into three parts. The first is the dataset reference name, e.g. IRE-like, the parameter values specified by the user and the starting time. The second is the predicted common structure element in the given family. The remaining of an output shows the structure element occurrences in each sequence in the paired parenthesis format.
RESULTS
GPRM is freely accessible at http://bioinfo.cis.nctu.edu.tw/service/gprm/. Corrections, suggestions and feedback should be sent to yhu@cis.nctu.edu.tw.
DISCUSSION
In this paper, we describe how to run GPRM properly. GPRM has a flexible RNA secondary structure representation. It is capable of dealing with structures more complex than simple stem-loops, such as pseudoknots.
GPRM is aimed at finding common secondary structure elements, not a global alignment, in a sufficiently large family (e.g. >15 members) of unaligned RNA sequences. It is not applicable to finding the possible folding of a single sequence. Besides, owing to the hardware limitation of our current PC server, GPRM is currently limited to finding structure elements with no more than five stems. This constraint is expected to be relaxed after we install GPRM on other higher-end computer platforms.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to the anonymous reviewers for their valuable comments and suggestions. Thanks also go to Chao-I Chen for improving the web-based I/O interface. This project was supported by National Science Council, Taiwan (NSC 91-2213-E-009-169).
REFERENCES
- 1.Gygi S.P., Rochon,Y., Franza,B.R. and Aebersold,R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol., 19, 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klaff P., Riesner,D. and Steger,G. (1996) RNA structure and the regulation of gene expression. Plant Mol. Biol., 32, 89–106. [DOI] [PubMed] [Google Scholar]
- 3.Gray N.K. and Hentze,M.W. (1994) Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep., 19, 195–200. [DOI] [PubMed] [Google Scholar]
- 4.Zuker M. and Stiegler,P. (1981) Optimal computer Folding of large RNA sequences using thermodynamic and auxiliary information. Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuker M. and Jacobson,A. (1995) Well-determined regions in RNA secondary structure prediction: analysis of small subunit ribosomal RNA. Nucleic Acids Res., 23, 2791–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gultyaev A.P., van Batenburg,F.H.D. and Pleij,C.W.A. (1995) The computer simulation of RNA folding pathways using a genetic algorithm. J. Mol. Biol., 250, 37–51. [DOI] [PubMed] [Google Scholar]
- 7.Rivas E. and Eddy,S.R. (1999) A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol., 285, 2053–2068. [DOI] [PubMed] [Google Scholar]
- 8.van Batenburg F.H.D., Gultyaev,A.P. and Pleij,C.W.A. (1995) An APL-programmed genetic algorithm for the prediction of RNA secondary structure. J. Theor. Biol., 174, 269–280. [DOI] [PubMed] [Google Scholar]
- 9.Eddy S. and Durbin,R. (1994) RNA sequence analysis using covariance models. Nucleic Acids Res., 22, 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J.-H., Le,S.-Y. and Maizel,J.V. (2000) Prediction of common secondary structures of RNAs: a genetic algorithm approach. Nucleic Acids Res., 28, 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorodkin J., Heyer,L.J. and Stormo,G.D. (1997) Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res., 25, 3724–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorodkin J., Stricklin,S.L. and Stormo,G.D. (2001) Discovering common stem-loop motifs in unaligned RNA sequences. Nucleic Acids Res., 29, 2135–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouthinon D. and Soldano,H. (1999) A new method to predict the consensus secondary structure of a set of unaligned RNA sequences. Bioinformatics, 15, 785–798. [DOI] [PubMed] [Google Scholar]
- 14.Hu Y. (2002) Prediction of consensus structural motifs in a family of coregulated RNA sequences. Nucleic Acids Res., 30, 3886–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonassen I., Collins,J.F. and Higgins,D.G. (1995) Finding flexible patterns in unaligned protein sequences. Protein Sci., 4, 1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]