Abstract
We present three programs for ab initio gene prediction in eukaryotes: Exonomy, Unveil and GlimmerM. Exonomy is a 23-state Generalized Hidden Markov Model (GHMM), Unveil is a 283-state standard Hidden Markov Model (HMM) and GlimmerM is a previously-described genefinder which utilizes decision trees and Interpolated Markov Models (IMMs). All three are readily re-trainable for new organisms and have been found to perform well compared to other genefinders. Results are presented for Arabidopsis thaliana. Cases have been found where each of the genefinders outperforms each of the others, demonstrating the collective value of this ensemble of genefinders. These programs are all accessible through webservers at http://www.tigr.org/software.
INTRODUCTION
Accurate gene structure prediction remains an important component of genomic annotation efforts, notwithstanding the increased availability, at least for some organisms, of hand-crafted annotations. Even though ab initio predictions are typically the least trusted form of evidence used by human annotators, they are necessary for ensuring high levels of sensitivity, in some cases being the only form of evidence for a gene that might otherwise be completely overlooked (1,2).
Unfortunately, genefinder programs for eukaryotes are far from perfect, often predicting genes or exons which do not exist, failing to predict those that do exist or generating predictions having one or more incorrect exon boundaries. Furthermore, different genefinders trained for the same organism often produce different predictions. Thus, while a genefinder is a useful component of the annotation process, it is difficult to choose a single genefinder to use and genome annotation systems often employ multiple genefinders in order to increase the number of predicted coding segments (CDSs).
We present here three genefinder programs, each with different strengths due to their different algorithmic designs. While all three programs achieved high levels of accuracy in our controlled tests, the programs sometimes do not agree on the structure of a given gene, with each of the programs occasionally producing a better prediction than the other two. For this reason, we believe that annotators will appreciate having access to all three programs for use in their annotation efforts.
All three programs can be run directly on genomic sequences by using the http interface at TIGR, available from http://www.tigr.org/software. This web interface will be useful to laboratories lacking the computational facilities for running our UNIX-based software themselves and to those working on small-scale projects, such as those for sequencing an individual BAC or specific region of a genome. Sequences <30 kb can be pasted directly into the browser, whereas larger sequences <200 kb can be uploaded from a user's system as a FASTA file. The resulting gene predictions can be displayed directly in the browser or emailed to the user. GlimmerM is also freely available for direct download, including source code that permits re-training the system on any species. (The user must collect training data to feed to the system.) Exonomy and Unveil will be made freely available as open source software in the near future.
The GlimmerM web server (http://www.tigr.org/tdb/glimmerm/glmr_form.html) can provide gene predictions for several organisms, including Plasmodium falciparum, P.yoelii, Oryza sativa (rice), Aspergillus fumigatus, Arabidopsis thaliana, Theileria parva and Schistosoma mansoni. The Exonomy and Unveil web servers, at http://www.tigr.org/tdb/Exonomy/exonomy.html and http://www.tigr.org/tdb/Unveil/unveil.html, respectively, were initially trained on A.thaliana, but other organisms will be added in the very near future and will probably be available at the time of this publication. On the web page, an organism can be selected using a drop-down list.
DESIGN AND IMPLEMENTATION
All three genefinders utilize statistical models trained from known genes to evaluate each gene structure. They employ dynamic programming techniques to search all possible gene structures in a DNA sequence and efficiently find the most probable structure, given the statistical model of the genefinder.
Each genefinder is based on one or more types of Markov model. The simplest is the nth-order Markov chain (MC), which calculates the conditional probability of a base, given the preceding n bases and then multiplies the conditional probabilities together to arrive at a probability for an entire subsequence. A variant of this scheme is the Interpolated Markov Model (IMM), which interpolates between models of different order, based on the amount of evidence available for a given probability estimate (3). Nonstationary Markov chains (NSMC) utilize two or more MCs in strict order while allowing the individual chains making up the NSMC to partition the sequence in any way to maximize the joint probability. Three-periodic Markov Chains (3PMC) utilize three MCs in a cycle, one for each frame within a putative coding region. Hidden Markov Models (HMM) are state-based models in which each state emits a single base with fixed probability and then transitions to another state with fixed probability. A Generalized Hidden Markov Model (GHMM) is an HMM in which each state can emit a sequence of bases rather than a single base and which allows explicit modeling of exon and intron lengths.
Models such as these, when used to evaluate variable-length DNA sequences, are referred to as content sensors. Other content sensors that we employ include codon bias models and decision trees. For fixed-length features such as splice sites and start/stop codons we employ several types of signal sensor, including Weight Matrices (WMM), also called Position Weight Matrices (PWM), Weight Array Matrices (WAM), Windowed Weight Array Matrices (WWAM), MCs and Maximal Dependence Decomposition (MDD) trees. Detailed treatments of content and signal sensors can be found in the literature (4–6).
Some general features of our three genefinders are as follows:
They can be configured to predict only complete gene structures or to allow partial genes.
They are retrainable on new organisms without needing to recompile the program source code.
They predict genes on either or both strands during a single pass over the DNA sequence.
They do not attempt to predict UTRs (untranslated regions), though some of them model UTRs internally.
They currently generate a single structure for each predicted gene (no alternative splicing capability).
GlimmerM
The basis of GlimmerM is a dynamic programming algorithm that considers all combinations of possible exons for inclusion in a gene model (by gene model we mean the protein coding portion of a gene or CDS) and chooses the best of all these combinations by using IMMs trained on complete coding regions (7). The decision about what gene model is best is a combination of the ‘strength’ of the splice sites and that of the potential coding regions, as we describe below.
The splice site predictor algorithm in GlimmerM (8) first uses MDD (9) and first-order MCs to capture dependencies among neighboring bases in a small window around each splice junction (16 and 29 bp around the donor and acceptor sites, respectively). The algorithm then takes advantage of the fact that the coding and non-coding sequences switch at the splice junction and tries to detect this switch with two second-order MCs, one which models coding sequence and another that models non-coding sequence. The length of each of these coding or non-coding context windows is currently fixed at 80 bp. By keeping only those splice sites having the maximal score within a 60 bp window, many false positives are eliminated.
Potential coding regions are evaluated by a scoring function based on decision trees that estimate the probability that a DNA subsequence is coding. Subsequences are evaluated according to their putative type: intron, initial exon, internal exon, final exon and single-exon gene. Each such subsequence is run through 10 different decision trees built with the OC1 system (10). The probabilities obtained with the decision trees are averaged to produce a smoothed estimate of the probability that the given subsequence is of a certain type.
A putative gene model is then accepted only if the IMM score for the coding sequence in the correct reading frame exceeds a fixed threshold.
Unveil
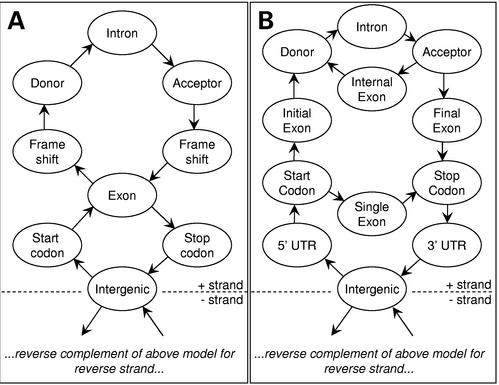
Unveil is a 283-state HMM based on the VEIL design (11). In our implementation the actual topology and number of states in the HMM are specified in a configuration file to afford some flexibility for future enhancements. When the model is loaded at run time, the program automatically reverse-complements this model to allow simultaneous prediction on the reverse strand. The overall structure currently in use is depicted schematically in Figure 1A. The 141 states comprising the forward-strand HMM are shown as being grouped into eight distinct submodels, each of which can be trained individually using the Baum–Welch EM algorithm (12) and automatically combined into a composite HMM. Thenceforth the HMM is treated as a single, complete model for the purpose of optimal path analysis.
Figure 1.
Simplified structure of two Markov model genefinders: (A) Unveil and (B) Exonomy.
Once the HMM and the subject sequence are loaded, the program generates a single set of non-overlapping gene structure predictions by applying the Viterbi algorithm (13) to find the most probable path through the states of the HMM that would emit the subject sequence, given the HMM's emission and transition probabilities. Bases emitted by coding states according to this optimal path are grouped into exons and similarly for intron states and introns, allowing the exons to then be grouped into genes. Coding frames are explicitly tracked to ensure that exons within a gene are in the same open reading frame. Viterbi decoding is achieved via an efficient dynamic programming implementation on a sparse graph.
The HMM currently in use was designed to model 5′ and 3′ UTRs, hexamer frequencies in introns, 7 bp consensus regions around splice sites, noncanonical start/stop codons and frame effects within exons. HMM states matching putative exons do not differentiate between the different exon types (e.g. initial, final).
The output format is GFF (General Feature Format, see http://www.sanger.ac.uk/Software/GFF).
Exonomy
Exonomy is a 23-state GHMM similar to Genscan (9) and Genie (14). The generalized nature of the GHMM framework provides greater flexibility over an HMM for tuning and augmenting the genefinder, because entire gene features such as exons and splice junctions are handled by individual states in the GHMM, which can be independently retrained, reparameterized, or replaced by other types of model to improve performance, without needing to recompile the program's source code.
For its content-sensor states Exonomy provides MCs, IMMs, 3PMCs, NSMCs and codon bias estimation. For its signal sensors it provides WMMs, WAMs, WWAMs, MCs and MDD trees having any of the former as leaf models. Splice sites and start and stop codons are identified using MDD trees and variable-length context sequences, as in GlimmerM. Currently, the overall state architecture of Exonomy is fixed into a 23-state topology which includes submodels for introns, intergenic segments, single-exon genes and initial-, internal- and final-exons (Fig. 1B).
Exonomy begins by identifying possible splice sites and start and stop codons. It then constructs a graph containing a vertex for each splice site or start/stop codon and an edge for each exon, intron or intergenic segment. Frame constraints are applied to reduce the size of the graph and then submodels specified in a configuration file are dynamically loaded and applied to evaluate the individual vertices and edges in the graph. A dynamic-programming algorithm is then used to rapidly find the highest-probability path, which corresponds to the optimal gene model. Exonomy's output format is GFF.
RESULTS AND DISCUSSION
Prediction accuracy of our genefinders was assessed by training and testing all three programs on a large set of full-length cDNAs from A.thaliana, all of which have been mapped to the genome (15). The programs were all trained on the same set of 8500 cDNAs and then tested on 300 sequences each containing one gene (also from the cDNA dataset) plus a margin of 100 bp on either side of the start and stop codons. This rather artificial test scenario is not intended to serve as a basis for a comprehensive comparison of genefinders but merely to demonstrate accuracy comparable to that of more established programs.
Predictions were scored based on nucleotide accuracy (how accurately each base was classified as coding versus noncoding); sensitivity and specificity of exons predicted exactly; and percentage of genes predicted exactly (i.e. consisting entirely of exactly predicted exons). We also provide results of running a version of Genscan trained for A.thaliana (16) on the same test set for comparison. Because we were not able to retrain Genscan on the training set used by the other programs, the comparison is imperfect. Results are shown in Table 1.
Table 1. Accuracy comparison among four genefinders for 300 genes based on full-length A.thaliana cDNAs. Genscan was not trained on the same training set.
| Program | Nucleotide accuracy (%) | Exon specificity (%) | Exon sensitivity (%) | Percentage of exact genes (%) |
|---|---|---|---|---|
| Unveil | 94 | 75 | 74 | 46 |
| Exonomy | 95 | 63 | 61 | 42 |
| GlimmerM | 93 | 71 | 71 | 44 |
| Genscan | 94 | 80 | 75 | 27 |
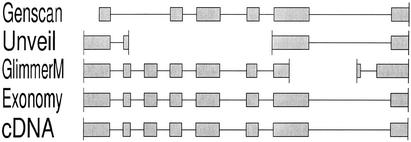
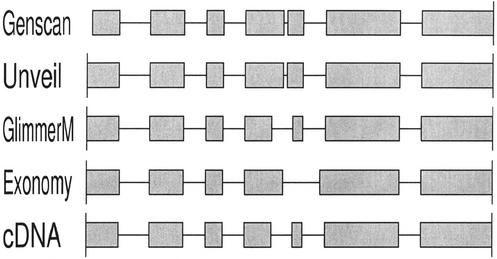
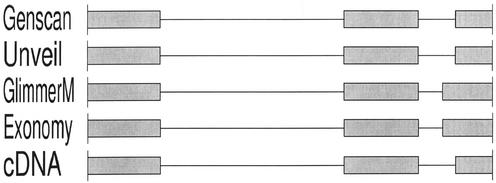
Table 1 shows that there is much room for improvement by all four genefinders. Not only was there variation in performance among the different programs, but the variation was different for each of the four measures of accuracy. In terms of numbers of perfectly correct gene predictions, Unveil exhibited the best performance and Genscan the worst, whereas according to exon sensitivity and specificity, Genscan performed the best, followed by Unveil. More importantly, the numbers show that all three of our genefinders perform quite respectably according to one or more measures of accuracy. More detailed comparison of the predictions on specific test sequences reveals that there are many cases of each of our genefinders outperforming the other three, as exemplified in Figures 2–4. Thus, were any of these genefinders to be excluded from an annotation system, overall ab initio prediction accuracy could be expected to suffer as a result.
Figure 2.
An example in which Exonomy produces the correct gene model.
Figure 4.
An example in which GlimmerM produces the correct gene model.
It should be noted that the four accuracy measures reported involve very different levels of discrimination, as the baseline nucleotide accuracy is expected to be very high for an organism with relatively low gene density, and as percentage of exact genes is very stringent (i.e. nearly-correct gene structures and wildly erroneous ones are all treated simply as errors). Furthermore, tradeoffs between these measures are possible, as evidenced by Genscan's high exon accuracy and rather lower gene accuracy, which situation can readily occur depending on the grouping of correct and incorrect exons. Nonetheless, Figures 2–4 demonstrate that imperfect gene predictions often are very near to being correct.
Note also that due to the possible existence of alternatively spliced forms in our test and training sets, we cannot be certain that some of those predictions which were classified as errors are not actually valid alternative transcripts, though this consideration should only strengthen the case for continuing to employ multiple genefinders, in addition to homology evidence, in any annotation project. Nevertheless, the need for additional improvements to genefinding techniques is clear. Our current research includes methods for combining genefinder predictions using various techniques, such as machine learning and multiobjective optimization, together with sequence homology and synteny information.
Figure 3.
An example in which Unveil produces the correct gene model (as does Genscan).
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by NSF grants MCB-0114792 and KDI-9980088, and by NIH grant R01-LM06845. The authors thank Jennifer Wortman, Mark Yandell and Art Delcher for helpful discussions and the anonymous reviewers for useful comments.
REFERENCES
- 1.Das M., Burge,C.B., Park,E., Colinas,J. and Pellitier,J. (2001) Assessment of the total number of human transcription units. Genomics, 77, 71–78. [DOI] [PubMed] [Google Scholar]
- 2.Guigo R., Dermitzakis,E.T., Agarwal,P., Ponting,C.P., Parra,G., Reymond,A., Abril,J.F., Keibler,E., Lyle,R., Ucla,C., Antonarakis,S.T. and Brent,M.R. (2003) Comparison of mouse and human genomes followed by experimental verification yields an estimated 1019 additional genes. Proc. Natl Acad. Sci. USA, 100, 1140–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salzberg S.L., Delcher,A.L., Kasif,S. and White,O. (1998) Microbial gene identification using interpolated Markov models. Nucleic Acids Res., 26, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salzberg S.L., Searls,D.B. and Kasif,S. (eds). (1998) Computational Methods in Molecular Biology. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 5.Durbin R., Eddy,S., Krogh,A. and Mitchison,G. (1998) Biological Sequence Analysis. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 6.Mathé C., Sagot,M.-F., Schiex,T. and Rouzé,P. (2002) Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res., 30, 4103–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salzberg S.L., Pertea,M., Delcher,A.L., Gardner,M.J. and Tettelin,H. (1999) Interpolated Markov models for eukaryotic gene finding. Genomics, 59, 24–31. [DOI] [PubMed] [Google Scholar]
- 8.Pertea M., Lin,X. and Salzberg,S.L. (2001) GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res., 29, 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 10.Murthy S.K., Kasif,S. and Salzberg,S.L. (1994) A system for induction of oblique decision trees. J. Artificial Intelligence Res., 2, 1–32. [Google Scholar]
- 11.Henderson J., Salzberg,S. and Fasman,K.H. (1997) Finding genes in DNA with a Hidden Markov Model. J. Comput. Biol., 4, 127–141. [DOI] [PubMed] [Google Scholar]
- 12.Baum L.E., Petrie,T., Soules,G. and Weiss,N. (1970) A maximization technique occurring in the statistical analysis of probabilistic functions of Markov chains. Ann. Math. Stat., 41, 164–171. [Google Scholar]
- 13.Viterbi A.J. (1967) Error bounds for convolutional codes and an asymptotically optimal decoding algorithm. IEEE Trans. Information Theory, 13, 260–269. [Google Scholar]
- 14.Reese M.G., Kulp,D., Tammana,H. and Haussler,D. (2000) Genie—gene finding in Drosophila melanogaster. Genome Res., 4, 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas B.J., Volfovsky,N., Town,C.D., Troukhan,M., Alexandrov,N., Feldmann,K.A., Flavell,R.B., White,O. and Salzberg,S.L. (2002) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol., 3, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]