Abstract
NCI is a server for the identification of non-canonical interactions in protein structures. These interactions, which include N-H···π, Cα-H···π, Cα-H···O=C and variants of them, were first observed in small molecules and subsequently in high-resolution protein structures. Such interactions have been subjected to extensive structural analysis to elucidate the different geometric criteria required to identify them. These interactions have also recently been shown to be important for the stability of protein structures. In this work, I describe a server called NCI, which allows the user to either upload protein/peptide coordinates in Protein Data Bank (PDB) format or enter a Structural Classification of Proteins database (SCOP)/PDB identifier for which NCI identifies the different non-canonical interactions, based purely on geometric criteria. Results are presented as an HTML table, as a parseable text file and as a color-coded interaction matrix. In addition, the user can view the RasMol image highlighting the interactions in the protein structure and download the RasMol script. The NCI server is available at: http://www.mrc-lmb.cam.ac.uk/genomes/nci/.
INTRODUCTION
A delicate balance between a variety of weak and strong non-covalent interactions contributes to the stability of proteins. Although hydrogen bonds (1–3), salt bridges (4,5) and hydrophobic interactions (6,7) are considered to be the major determinants of structural stability, in recent years non-canonical interactions have been shown to be of much greater importance than previously thought, particularly those interactions in which the π ring system serves as a hydrogen bond acceptor.
These non-canonical interactions involving the π ring system as hydrogen bond acceptor were first described by Wulf et al. (8) through spectroscopic analysis of small molecules and subsequently in peptides by McPhain and Sim (9). The occurrence of Cα-H···O=C hydrogen bonds were documented by Sutor (10) and later studied in great detail by Desiraju and Steiner (11). Even though these non-canonical interactions were discovered a long time ago, their importance was not immediately appreciated. Only in recent years have they been implicated to serve as an additional stabilizing factor in beta sheets (12), helix termini (13), helices containing proline residues (14), packing of transmembrane helices (15), collagen (16) and DNA (17).
Further investigations by various research groups have established the role of non-canonical interactions in a variety of functions such as ligand recognition (18), DNA recognition (19), enzymatic action (20), stabilization of secondary structures (21) and protein–protein complexes (22). Theoretical ab initio calculations have also been performed (23–26) and have shown that the energy of these non-canonical interactions is less than the energy of a conventional hydrogen bond. However, since these interactions can occur more frequently than regular hydrogen bonds, they may well contribute to the protein's stability to the same extent as standard hydrogen bonds (22).
With the availability of a number of high-resolution structures in the Protein Data Bank (PDB), there have been large-scale studies performed on specific interactions to get insight into their prevalence in protein structures and to establish the geometric criteria required to identify them. Recently, Steiner and Koellner (27) have performed a comprehensive survey on the occurrence of such non-canonical hydrogen bonds involving π acceptors in proteins and analysed recurrent structural patterns involving these interactions. Other studies by Derewenda et al. (28), Gallivan and Dougherty (29), Brandl et al. (30) and Toth et al. (31) provide a similar insight into the occurrence of Cα-H···O=C, cation-π and Cα-H···π interactions in protein structures.
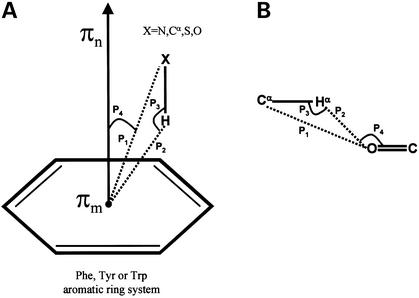
In this article, I describe a tool called NCI which uses previously published geometric criteria to identify these non-canonical interactions for a given PDB (32) or Structural Classification of Proteins database (SCOP) domain (33,34) coordinate file. It makes sense to calculate non-canonical interactions for structures solved at 2.5 Å resolution or better. Figure 1 illustrates the geometric parameters that are commonly used to identify such interactions.
Figure 1.
Geometric criteria used to identify non-canonical interactions. (A) The aromatic ring system is represented as a hexagon. In the case of Trp, two ring systems, a five-member and a six-member ring system, are considered separately. The donor group is represented as X-H, where X can be main chain Cα, N atom or side chain N atom of Arg, Lys, S atom of Cys and O atom of Thr, Ser and Tyr. πm represents the ring mid-point and the vector πn represents the normal to the plane of the ring. P1 and P2 are distances from X and H to πm respectively. P3 is the angle between vectors X-H and H-πm and P4 is the angle between the vectors X-πm and the ring normal, πn. (B) Parameters used to identify Cα-H···O=C interactions. P1 and P2 are distances from Cα, Hα to the main chain carbonyl O. P3 and P4 are the angles C-H···O and H···O=C, respectively. Default values used to identify interactions based on these definitions are provided in Table 1.
INPUT DATA AND NCI PARAMETERS
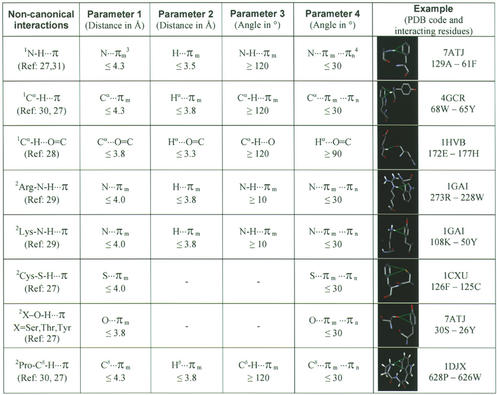
Input to the NCI server is either: (i) a PDB identifier; (ii) a SCOP domain identifier; or (iii) an uploaded peptide/protein coordinate file in PDB format. NCI provides the option of identifying up to eight types of non-canonical interactions in the current version. These include three main chain–side chain interactions (N-H···π, Cα-H···π and Cα-H···O=C) and five side chain–side chain interactions [Arg-N-H···π, Lys-N-H···π, Pro-Cδ-H···π, Cys-S-H···π and (Ser, Thr, Tyr)-O-H···π]. Each interaction can be identified according to four geometric criteria and the user has the option of adjusting these parameters. Default values are shown in Table 1 which also includes references to articles in which individual interactions are described in full detail.
Table 1. Default parameters used to calculate non-canonical interactions by NCI.
OUTPUT FROM NCI
The output of the NCI server is available in four different formats:
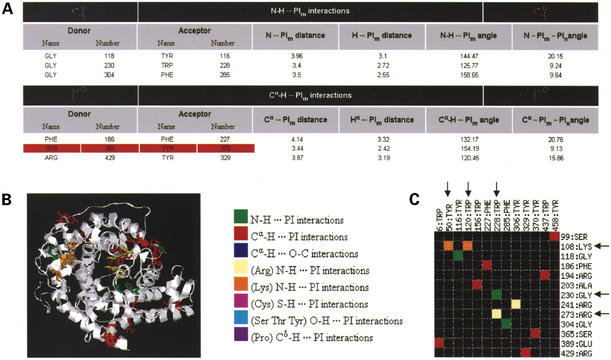
An HTML table (Fig. 2A) reporting all the interactions and the observed values for each of the parameters. Contacts between badly positioned residues from low resolution regions in the structure are colored red.
A parseable text file, which can be downloaded for further analysis, for example to identify interactions that occur at a protein–protein interface.
A RasMol (35) image (Fig. 2B) in which the protein is displayed in cartoon representation and residues involved in specific interactions are colored differently and displayed in ‘stick’ representation. Both the RasMol script and the file including coordinates of the added hydrogen atoms can be downloaded for further analysis.
A schematic interaction matrix (Fig. 2C) that represents the interactions in a color-coded form. This can be used to visualize the results at a glance. It also provides a means to immediately identify residues that are involved in multiple interactions.
Figure 2.
Sample output for glucoamylase (PDB code: 1GAI, 1.7 Å resolution structure). (A) The HTML table provides values for the different parameters for each type of non-canonical interactions observed in the structure. A red background is indicative of bad contacts in the structure. (B) An image generated by the server using RasMol. It highlights the different non-canonical interactions in the structure according to the color code given in the key for (B) and (C). Residues involved in a non-canonical interaction are displayed in stick representation. Users can download a PS or a PNG file of the image as well as the RasMol script used to generate the image. (C) A schematic interaction matrix provides a quick way of identifying residues involved in multiple interactions. Acceptor residues are shown on the X-axis and donor residues on the Y-axis. In this case, we can see that Lys:108 is involved simultaneously in two N-H···π interactions with Tyr:50 and Trp:120. Similarly, Trp:228 is involved in two different types of interactions, a main chain N-H···π interaction with Gly:230 and a side chain N-H···π interaction with Arg:273.
IMPLEMENTATION AND ORGANIZATION
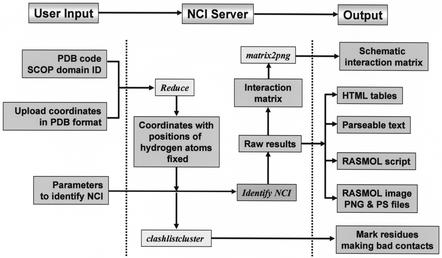
The program to compute non-canonical interactions uses atom information and coordinate data in the structure file to calculate various distance and angle parameters (Fig. 1 and Table 1), based on the values for the parameters as chosen by the user. The program is written in PERL and the web interface has been implemented using CGI-PERL. The NCI server also makes use of two previously published programs called ‘REDUCE’ (36) (to fix the positions of hydrogen atoms in the coordinate file) and ‘matrix2png’ (37) (to create the color-coded interaction matrix). The program also marks residues that make bad contacts in the structure (due to poor refinement or disordered regions) according to the output of the REDUCE/clashlistcluster (36) program. Additionally, the NCI results for the 12 neutron structures available in the PDB (as of 7 March 2003) are available on the website, mainly as reference and as indication of what can be considered a non-canonical interaction for structures in which the hydrogen atom coordinates are experimentally determined. The organization of the NCI server is shown in Figure 3.
Figure 3.
An example in which Unveil produces the correct gene model (as does Genscan).
CONCLUSIONS AND FUTURE DIRECTIONS
Non-canonical interactions play an important role in stabilizing protein structures and protein–protein interfaces. The NCI server is a useful tool for identifying such interactions in old and new structures. Its results can be used for a variety of purposes, ranging from rational design of mutagenesis experiments to the analysis of conservation of interactions in protein families and at functional sites. Future additions to the server will include identification of non-canonical interactions at protein–DNA and protein–ligand interfaces, pre-computed results for all SCOP domains and PDB structures and the possibility to perform large-scale analyses on related proteins.
Acknowledgments
ACKNOWLEDGEMENTS
I am grateful to Dr Loredana Lo Conte for stimulating and knowledgeable discussions and for correcting the manuscript. I would also like to acknowledge my supervisor Dr Sarah Teichmann for reading the manuscript and for her encouragement; Raj, Daniel and Murali for their comments on the website; and Professor Balaram for introducing me to the field of NCI. I would like to thank the anonymous referees for valuable suggestions. I am grateful to the Medical Research Council, Cambridge Commonwealth Trust and Trinity College, Cambridge, for financial support.
REFERENCES
- 1.Baker E.N. and Hubbard,R.E. (1984) Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol., 44, 97–179. [DOI] [PubMed] [Google Scholar]
- 2.Jeffrey G.A. and Saenger,W. (1994) Hydrogen Bonding in Biological Structures. Springer Verlag, New York, NY. [Google Scholar]
- 3.Creighton T. (1993) Proteins: Structures and Molecular Properties, 2nd edn. W.H. Freeman and Co., New York. [Google Scholar]
- 4.Horovitz A., Serrano,L., Avron,B., Bycroft,M. and Fersht,A. (1990) Strength and co-operativity of contributions of surface salt bridges to protein stability. J. Mol. Biol., 216, 1031–1044. [DOI] [PubMed] [Google Scholar]
- 5.Pace C.N., Shirley,B.A., McNutt,M. and Gajiwala,K. (1996) Forces contributing to the conformational stability of proteins. FASEB J., 10, 75–83. [DOI] [PubMed] [Google Scholar]
- 6.Dill K.A. (1990) Dominant forces in protein folding. Biochemistry, 29, 7133–7155. [DOI] [PubMed] [Google Scholar]
- 7.Lins L. and Brasseur,R. (1995) The hydrophobic effect in protein folding. FASEB J., 9, 535–540. [DOI] [PubMed] [Google Scholar]
- 8.Wulf O.R., Liddel,U. and Hendricks,S.B. (1936) The effect of ortho substitution on the absorption of the OH group of phenol in the infrared. J. Am. Chem. Soc., 58, 2287–2293. [Google Scholar]
- 9.McPhail A.T. and Sim,G.A. (1965) Hydroxyl-benzene hydrogen bonding. An X-ray study. Chem. Commun., 124–125. [Google Scholar]
- 10.Sutor D.J. (1962) The C-H···O hydrogen bonds in crystals. Nature, 195, 68–69. [Google Scholar]
- 11.Desiraju G.R. and Steiner,T. (1999) The Weak Hydrogen Bond in Structural Chemistry and Biology. Oxford University Press, Oxford. [Google Scholar]
- 12.Fabiola G.F., Krishnaswamy,S., Nagarajan,V. and Pattabhi,V. (1997) C-H···O hydrogen bonds in beta sheets. Acta Crystallog. Sect. D., 53, 316–320. [DOI] [PubMed] [Google Scholar]
- 13.Madan Babu M., Kumar Singh,S. and Balaram,P. (2002) A C-H···O hydrogen bond stabilized polypeptide chain reversal motif at the C terminus of helices in proteins. J. Mol. Biol., 322, 871–880. [DOI] [PubMed] [Google Scholar]
- 14.Chakrabarti P. and Chakrabarti,S. (1998) C-H···O hydrogen bond involving proline residues in alpha-helices. J. Mol. Biol., 284, 867–873. [DOI] [PubMed] [Google Scholar]
- 15.Senes A., Ubarretxena-Belandia,I. and Engelman,D.M. (2001) The C-H···O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl Acad. Sci. USA, 98, 9056–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bella J. and Berman,H.M. (1996) Crystallographic evidence for C-H···O=C hydrogen bonds in a collagen triple helix. J. Mol. Biol., 264, 734–742. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh A. and Bansal,M. (1999) C-H·O hydrogen bonds in minor groove of A-tracts in DNA double helices. J. Mol. Biol., 294, 1149–1158. [DOI] [PubMed] [Google Scholar]
- 18.Kryger G., Silman,I. and Sussman,J.L. (1999) Structure of acetylcholinesterase complexed with E2020: implications for the design of new anti-alzheimer drugs. Structure, 7, 297–307. [DOI] [PubMed] [Google Scholar]
- 19.Parkinson G., Gunasekera,A., Vojtechovsky,J., Zhang,X., Kunkel,T.A., Berman,H. and Ebright,R.H. (1996) Aromatic hydrogen bond in sequence-specific protein DNA recognition. Nature Struct. Biol., 3, 837–841. [DOI] [PubMed] [Google Scholar]
- 20.Derewenda Z.S., Derewenda,U. and Kobos,P.M. (1994) (His)Cɛ-H···O=C hydrogen bond in the active sites of serine hydrolases. J. Mol. Biol., 241, 83–93. [DOI] [PubMed] [Google Scholar]
- 21.Armstrong K.M., Fairman,R. and Baldwin,R.L. (1993) The (i, i+4) Phe-His interaction studied in an alanine-based alpha-helix. J. Mol. Biol., 230, 284–291. [DOI] [PubMed] [Google Scholar]
- 22.Jiang L. and Lai,L. (2002) C-H···O hydrogen bonds at protein-protein interfaces. J. Biol. Chem., 277, 37732–37740. [DOI] [PubMed] [Google Scholar]
- 23.Scheiner S., Kar,T. and Gu,Y. (2001) Strength of the C-H···O hydrogen bond of amino acid residues. J. Biol. Chem., 276, 9832–9837. [DOI] [PubMed] [Google Scholar]
- 24.Vargas R., Garza,J., Dixon,D.A. and Hay,B.P. (2000) How strong is the C-H···O=C hydrogen bond? J. Am. Chem. Soc., 122, 4750–4755. [Google Scholar]
- 25.Levitt M. and Perutz,M.F. (1988) Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol., 201, 751–754. [DOI] [PubMed] [Google Scholar]
- 26.Duan G., Smith,V.H.Jr. and Weaver,D.F. (1999) An ab initio and data mining study on aromatic-amide interactions. Chem. Phys. Lett., 310, 323–332. [Google Scholar]
- 27.Steiner T. and Koellner,G. (2001) Hydrogen bonds with pi-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J. Mol. Biol., 305, 535–557. [DOI] [PubMed] [Google Scholar]
- 28.Derewenda Z.S., Lee,L. and Derewenda,U. (1995) The occurrence of C-H···O hydrogen bonds in proteins. J. Mol. Biol., 252, 248–262. [DOI] [PubMed] [Google Scholar]
- 29.Gallivan J.P. and Dougherty,D.A. (1999) Cation-pi interactions in structural biology. Proc. Natl Acad. Sci. USA, 96, 9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandl M., Weiss,M.S., Jabs,A., Suhnel,J. and Hilgenfeld,R. (2001) C-H···PI-interactions in proteins. J. Mol. Biol., 307, 357–377. [DOI] [PubMed] [Google Scholar]
- 31.Toth G., Watts,C.R., Murphy,R.F. and Lovas,S. (2001) Significance of aromatic-backbone amide interactions in protein structure. Proteins, 43, 373–381. [DOI] [PubMed] [Google Scholar]
- 32.Westbrook J., Feng,Z., Chen,L., Yang,H. and Berman,H.M. (2003) The Protein Data Bank and structural genomics. Nucleic Acids Res., 31, 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo Conte L., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandonia J.M., Walker,N.S., Lo Conte,L., Koehl,P., Levitt,M., Brenner,S.E. (2002) ASTRAL compendium enhancements. Nucleic Acids Res., 30, 260–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sayle R. and Milner-White,E.J. (1995) RasMol: Biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]
- 36.Word J.M., Lovell,S.C., Richardson,J.S. and Richardson,D.C. (1999) Asparagine and glutamine: using hydrogen atom contacts in the choice of sidechain amide orientation. J. Mol. Biol., 285, 1735–1747. [DOI] [PubMed] [Google Scholar]
- 37.Pavlidis P. and Noble,W.S. (2003) Matrix2png: a utility for visualizing matrix data. Bioinformatics, 19, 295–296. [DOI] [PubMed] [Google Scholar]