Abstract
The World Wide Web server of the PBIL (Pôle Bioinformatique Lyonnais) provides on-line access to sequence databanks and to many tools of nucleic acid and protein sequence analyses. This server allows to query nucleotide sequence banks in the EMBL and GenBank formats and protein sequence banks in the SWISS-PROT and PIR formats. The query engine on which our data bank access is based is the ACNUC system. It allows the possibility to build complex queries to access functional zones of biological interest and to retrieve large sequence sets. Of special interest are the unique features provided by this system to query the data banks of gene families developed at the PBIL. The server also provides access to a wide range of sequence analysis methods: similarity search programs, multiple alignments, protein structure prediction and multivariate statistics. An originality of this server is the integration of these two aspects: sequence retrieval and sequence analysis. Indeed, thanks to the introduction of re-usable lists, it is possible to perform treatments on large sets of data. The PBIL server can be reached at: http://pbil.univ-lyon1.fr.
INTRODUCTION
Since the explosion of the web, biologists no longer need to have accounts on centralized servers or to use email for accessing sequence collections or to run analysis programs. With a connection to the Internet and a web browser it is possible to access easily almost all existing data banks and software devoted to sequence study. Some limitations still exist in the use of such servers. A significant problem is that it is often not possible to keep track of the previous interrogations from one session to another: once the user is disconnected from the server, all his/her work is lost. Also, there are no real links between the two aspects of sequence retrieval and sequence analysis. Usually, a method can be launched only on a single sequence and not on a set of sequences previously selected by the user. To fill in these lacunas, we have developed a query system that integrates the notion of re-usable sequence lists, as well as tools for performing treatments on these lists. The major general sequence data banks as well as locally developed, more specialized systems, can be accessed through the PBIL server. Among the functionalities integrated are: similarity search programs, multiple alignment methods, simple statistics on sequences, protein secondary structure prediction, molecular modelling and a complete set of multivariate methods. All these tools are set up as CGI (Common Gateway Interface) programs written in C or Perl and their results are sent as simple HTML documents.
SYSTEM AND METHODS
Databanks access
The first layer of the system is represented by a set of databanks indexed with the ACNUC system (1). Presently, there are 11 nucleotide or protein databases accessible at PBIL and their list is given in Table 1. Any collection in the EMBL, GenBank, SWISS-PROT or PIR formats can be indexed with ACNUC and installed on the server. The ACNUC structure allows the use of several criteria to retrieve sequences, including sequence names, accession numbers, keywords, taxonomic data, bibliographic references, date of insertion in the bank and result of previous selection operation. Some criteria, like keywords, authors' names and sequence names accept the use of wildcards. Up to four criteria can be combined with logical operators. ACNUC also integrates the useful notion of subsequence: each kind of genomic fragment described in the features (e.g. CDS, tRNA or rRNA) can be defined as a subsequence and retrieved independently from its parent sequence. After a selection is made, the user has the possibility to retrieve sequences in different formats, this either by directly sending to the Web browser or by FTP access (recommended for large numbers of sequences).
Table 1. List of the sequence databanks accessible from the PBIL server.
| Name | Content | Ref. |
|---|---|---|
| GenBank | General (nucleotide) | (2) |
| EMBL | General (nucleotide) | (3) |
| SWISS-PROT+TrEMBL | General (protein) | (4) |
| PIR | General (protein) | (5) |
| HOVERGEN | Homologous vertebrate genes (nucleotide+protein) | (6) |
| HOBACGEN | Homologous bacterial genes (nucleotide+protein) | (7) |
| NUREBASE | Nuclear receptors in metazoa (nucleotide+protein) | (8) |
| RTKdb | Tyrosine kinase receptors (nucleotide+protein) | (9) |
| EMGLib | Prokaryotic complete genomes (nucleotide) | (10) |
| NRSub | Bacillus subtilis annotated genome (nucleotide) | (11) |
| NPSA_3DSeq | PDB entries at different levels of redundancy | — |
In the case of the homologous gene databanks developed at PBIL (HOBACGEN, HOVERGEN, NUREBASE and RTKdb), the information carried by the organization in families is very important and so requires specific tools to handle it. For that purpose, we have developed a dedicated interface: FamFetch (7). One problem is that this method of accessing these banks could be inconvenient for an occasional use and depends on the computer abilities of the user. This is why we have implemented on the PBIL server, a set of programs allowing to perform queries centred on families rather than on sequence entries. For instance, it is possible to retrieve all gene families that are shared by a given set of taxa and that are not present in a second set of taxa. Any taxonomic level can be used and mixed to compose the query (e.g. Homo sapiens, Primate, Mammalia). The first set of taxa can be used for an ‘inclusive’ or an ‘exclusive’ selection of families. With the inclusive search, any family presenting at least one sequence of each taxa of the list will be selected and with the exclusive search, the families presenting exclusively taxa of the list will be selected. Moreover families can be pre-selected according to the number of sequences and/or the number of species. This is useful to avoid families presenting only one sequence or species.
Each time a set of sequences or families is built, the corresponding list is stored on the server for a period of 24 h. During that time, it is available to the user either for sequence retrieval, query composition or use with sequence analysis tools.
The databanks of gene families also include alignments and trees for each family. Different possibilities are provided to represent and handle these two peculiar kinds of data, we provide different possibilities. First, alignments can be simply displayed with a colouring scheme in an HTML document or can be viewed with the JalView applet (http://www.ebi.ac.uk/~michele/jalview/contents.html). Trees can be displayed with the ATV (A Tree Viewer) applet (12). Second, it is possible to use helper applications as specific MIME-types were defined for these documents when we developed the first version of the server 7 years ago (13). A recommended helper for the alignments is SeaView (14), while NJplot (13) is a good option to visualize phylogenetic trees. Lastly, it is possible to store the corresponding files locally and to visualize them with ad hoc programs. The format used is Clustal for the alignments and NEWICK for the trees.
Simple sequence analysis
Many simple programs for nucleotide or protein sequence analyses are available for on-line use at PBIL. Pattern search utilities, computation of codon usage indexes, CDS (Coding DNA Sequences) prediction, local pairwise alignments, contigs assembly and CpG islands promoter detection (15) are among the tools provided for nucleotide sequences study. In the case of codon usage analyses, different indexes are available for use and it is even possible to compute the dedicated reference tables that are required by an index such as CAI (Codon Adaptation Index) (16). The PBIL also provides original methods (PATTINPROT and PROSCAN) to scan a protein sequence or a protein database for one or several pattern(s) using the PROSITE syntax. These programs use a weigthing function to ponderate the mismatch as a function of biological relevance. Signatures detection by using the InterPro (17) database is also available. Miscellanous tools are also present to locate helix-turn-helix DNA binding motif, antigenic regions, hydrophobic streches and transmembranous helices.
Similarity searches
Similarity searches can be performed by BLAST, PSI-BLAST (19), SSEARCH (20) and FASTA (21) on several nucleotide or protein sequence databanks or subsets of them obtained from ACNUC or SRS (18) queries (see Table 2 for a complete listing of the available banks and of the related programs). After a similarity search, the user may build a personal sequence databank with full or partial sequences or alternatively filter the output using different criteria: taxonomic data, keywords or date of insertion in the databank. An HTML interface has been developed to graphically display similarity search results. It provides three links for each subject sequence which allow one to quickly find relevant biological information by: (i) retrieving the database entry; (ii) checking the corresponding alignment from the similarity search results; and (iii) analyzing the subject sequence. This last point is provided by a link that permits the use of various biocomputing tools on the given sequence or to send a query to Geno3D, which is a service dedicated to molecular modelling of three-dimensional (3D) protein structures (22). If the subject sequence comes from a databank of known 3D structures (NPSA_3DSeq), additional functionalities allow the user to retrieve 3D data from other servers [such as SCOP (23), CATH (24) or PDB (25)] or to display the structure on a viewer software such as RasMol.
Table 2. List of programs and databanks available for similarity search on the PBIL server.
| Sequences |
Program |
Databank |
| Nucleotide | BLAST | GenBank without ESTs, GSSs, HTGs |
| GenBank ESTs | ||
| GenBank GSSs | ||
| GenBank HTGs | ||
| rRNAs | ||
| EMGLib | ||
| ACUTS | ||
| Human ESTs | ||
| Human GSSs | ||
| Human HTGs | ||
| Human without ESTs, GSSs, HTGs | ||
| Human DNA+cDNA | ||
| Protein | BLAST/PSI-BLAST | NR |
| SWISS-PROT | ||
| SWISS-PROT+TrEMBL | ||
| HOBACGEN | ||
| PDB | ||
| GenBank translation | ||
| EMGLib translation | ||
| Human | ||
| Yeast | ||
| Bacillus subtilis | ||
| Drosophila melanogaster | ||
| Caenorhabditis elegans | ||
| Arabidopsis thaliana | ||
| FASTA/SSEARCH | SWISS-PROT | |
| PDB | ||
| HMMER | SWISS-PROT | |
| PDB |
Multiple alignments
Multiple alignments of protein sequences can be performed by using a parallel version of ClustalW (26) or MULTALIN (27). Several choices are offered in the results page. For example, the alignment width can be modified as well as the multiple alignment display mode (all or only identical/conserved/different residues). A colour-coding scheme depending upon the identity level is available and a primary consensus is calculated on the fly. Moreover, this multiple alignment is fully coupled with protein secondary structure prediction (either isolated methods or deduced consensus), SOV (Structural OVerlap) compatibility calculation (28) can be used as a starting point to build a HMM (Hidden Markov Model) based profile with the HMMER package (29). The interactive edition of alignments can be performed by helper programs such as ANTHEPROT (30) or MPSA (31). These two items of software are also able to send analysis [e.g. SOPMA (32), PHD (33)] to the server thanks to client/server capabilities.
Protein structure prediction
Twelve methods for protein secondary structure prediction are available on the server as well as a dynamic computation of their consensus. These secondary structures can be checked for their compatibility (34) in order to validate (in the 10–30% identity range) the templates identified after a PSI-BLAST run. Then, 3D constraints are calculated from the selected template(s) and are used to build a molecular model of the query sequence. To validate the modelling process we provide: (i) a matrix of pairwise root mean square deviations (RMSD) computed from structural superimpositions of the models and the template(s) with associated plots; (ii) PROCHECK (35) outputs for geometric and stereochemical analysis; and (iii) a report of models energy. All files generated during the modelling process are stored in an archive that can be downloaded from the Geno3D server. The URL of that archive is sent by email to the user when the modelling is completed.
Multivariate statistics
The multivariate statistics methods integrated in the server come from the ADE-4 package (36). As this package has been ported under R, a freely available language and environment for statistical computing and graphics (http://www.r-project.org/), and as a web interface to R (http://www.math.montana.edu/Rweb/Resources.html) has been installed on the server, it is possible to use all ADE-4 methods for sequence study. The methods working on absolute frequencies or on distance matrices are particularly interesting, because frequencies of codons or amino acids can be computed from a set of selected sequences. The methods that can be applied then are, for example, CA (Correspondence Analysis), PCA (Principal Component Analysis) or DCA (Discriminant Correspondent Analysis) (37). Euclidean distance matrices can be computed on a set of multiply aligned sequences and this matrix can then be analyzed with different methods, such as PCO (Principal COordinates analysis) (13) or other methods available in R, such as cluster analysis. Many other multivariate analysis methods adapted to frequency tables and distance matrices are available in ADE-4, but have not yet been explored. Note that the complete ADE-4 documentation in HTML format can be accessed through our server. This documentation includes commented examples that can be used directly through copy/paste operations of the R commands.
EXAMPLES OF USE
Nucleotide sequence analysis
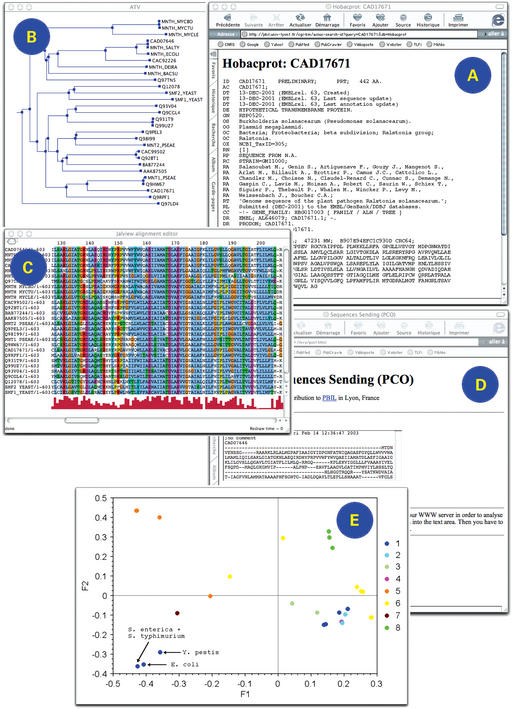
An example of nucleotide sequence study using some of the resources available from the server is given in Figure 1. Here we have studied the HOBACGEN family HBG017003 which corresponds to a manganese transport protein in bacteria. From the hypertext links associated with the family number (Fig. 1A), it is possible to access the list of all sequences belonging to the family, but also to the corresponding tree (Fig. 1B) and alignment (Fig. 1C). On the tree, we can see that the sequences (CAD07646, MNTH_SALTY, MNTH_ECOLI, CAC92226) of four Enterobacteria (Salmonella enterica, Salmonella typhimurium, Escherichia coli and Yersinia pestis, respectively) are not grouped with the other Proteobacteria. After retrieving the multiple alignment of the family, we submitted it to the form allowing to compute a distance matrix between the sequences (Fig. 1D). Once the matrix has been calculated, we ran a PCO on it with R. On the plot drawn by crossing the two first axes of the analysis, the sequences from the four Enterobacteria are clearly separated from the other Proteobacteria (Fig. 1E). This phenomenon could be explained either by a higher evolutionary rate for this gene in the clade of Enterobacteria or by a horizontal transfer in the common ancestor of the four species considered.
Figure 1.
Example of nucleotide sequence analysis at PBIL. (A) Family identification. (B) Tree visualization. (C) Alignment visualization. (D) Alignment submission for distance matrix computation. (E) PCO plot visualization 1 (blue) gamma proteobacteria; 2 (turquoise) alpha proteobacteria; 3 (light green) beta proteobacteria; 4 (purple) cyanobacteria; 5 (orange) high-G+C gram positives; 6 (yellow) low-G+C gram positives; 7 (brown) Thermus/Deinococcus group; 8 (green) eucarya.
Protein structure prediction
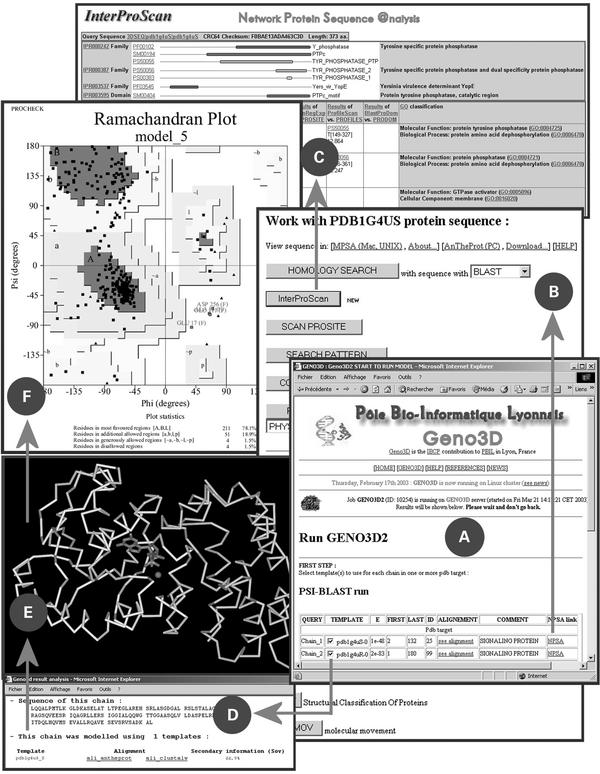
The typical work flow of the Geno3D tool designed for molecular modelling is shown in Figure 2. The first step consists in performing a PSI-BLAST run with the query sequence on a non-redundant databank (NR). From the list of hits displayed in the main result page (Fig. 2A), there are two links and one checkbox per hit. The first link is to analyze the given sequence with the methods available in NPS@ (38) through the NPSA link page (Fig. 2B). For example, the result of an InterProScan is shown in Figure 2C. The second link is to display the pairwise alignment. The checkbox is useful to select the corresponding template(s). In the case of low identity (Fig. 2D) between the query and the template, the secondary structure prediction (66.9%) is used to validate the template [for details, see (34)]. Spatial restraints are measured on the template structure and applied to the sequence to be modelled by using the CNS program (39). Once the models are generated (a dimer in Fig. 2E), the user receives a mail with a link to an archive containing the model and the modelling report (Fig. 2F).
Figure 2.
Typical flow chart for a molecular modelling at PBIL. (A) PSI-BLAST result page for query sequence. (B) NPSA link. (C) InterProScan result page. (D) Template choice and validation. (E) Model of a dimer (with a ligand). (F) PROCHECK analysis of the model.
DISCUSSION AND CONCLUSION
The PBIL server provides convenient and flexible tools for selecting sequences and then to analyze them through the web. It is widely used by the international scientific community as testified by usage statististics available at http://pbil.univ-lyon1.fr/usage/ (nucleotide sequences analysis and query) and http://npsa-pbil.ibcp.fr/cgi-bin/npsa_viewcnt.pl (protein structures). Classical features of other on-line sequence databanks browsers like SRS or Entrez (40) are integrated: possibility to build multicriteria queries, implementation of cross-references between different databanks as hypertext links, access to the parent sequence and to the different fragments of biological interest, option to transfer to the client all the sequences belonging to a list, etc. It provides an original feature that is not available in other systems: the link between sequence retrieval and sequence analysis tools.
In order to cope with ‘high throughput biology’ (complete genome sequencing and structural genomics), the automatization of the bioinformatic treatments is a crucial requirement to perform large scale comparative analysis. The constant updates of algorithms, the addition of new methods and databases are priorities for the PBIL. This requires powerful infrastructures with high storage and computational capacities. Data GRID approaches constitute a probable solution for such requirements. However, some problems still remain to be solved such as the distribution of up-to-date databases and the adaptation of bioinformatic algorithms to the GRID architecture.
Acknowledgments
ACKNOWLEDGEMENTS
S.P. is recipient of a fellowship from the EEC under the specific RTD program ‘Quality of Life and Management of Living Resources’, contract number QLRI-CT-2001-00015 for TEMBLOR. C.C. is recipient of a fellowship from ANRS. The PBIL is supported by CNRS, MENRT, INRIA, Région Rhône-Alpes and Génopole Rhône-Alpes.
REFERENCES
- 1.Gouy M., Gautier,C., Attimonelli,M., Lanave,C. and di Paola,G. (1985) ACNUC—a portable retrieval system for nucleic acid sequence databases: logical and physical designs and usage. Comput. Appl. Biosci., 1, 167–172. [DOI] [PubMed] [Google Scholar]
- 2.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stoesser G., Baker,W., van den Broek,A., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R. et al. (2003) The EMBL Nucleotide Sequence Database: major new developments. Nucleic Acids Res., 31, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeckmann B., Bairoch,A., Apweiler,R., Blatter,M.C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C.H., Yeh,L.S.L., Huang,H., Arminski,L., Castro-Alvear,J., Chen,Y., Hu,Z., Kourtesis,P., Ledley,R.S., Suzek,B.E. et al. (2003) The Protein Information Resource. Nucleic Acids Res., 31, 345–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duret L., Perrière,G. and Gouy,M. (1999) HOVERGEN: database and software for comparative analysis of homologous vertebrate genes. In Letovsky,S. (ed.), Bioinformatics Databases and Systems. Kluwer Academic Publishers, Boston, pp. 13–29. [Google Scholar]
- 7.Perrière G., Duret,L. and Gouy,M. (2000) HOBACGEN: database system for comparative genomics in bacteria. Genome Res., 10, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duarte J., Perrière,G., Laudet,V. and Robinson-Rechavi,M. (2002) NUREBASE: database of nuclear hormone receptors. Nucleic Acids Res., 30, 364–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grassot J., Mouchiroud,G. and Perrière,G. (2003) RTKdb: database of receptor tyrosine kinase. Nucleic Acids Res., 31, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrière G., Bessières,P. and Labedan,B. (2000) EMGLib: the Enhanced Microbial Genomes Library (update 2000). Nucleic Acids Res., 28, 68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrière G., Gouy,M. and Gojobori,T. (1998) The non-redundant Bacillus subtilis (NRSub) database: update 1998. Nucleic Acids Res., 26, 61–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zmasek C.M. and Eddy,S.R. (2001) ATV: display and manipulation of annotated phylogenetic trees. Bioinformatics, 17, 383–384. [DOI] [PubMed] [Google Scholar]
- 13.Perrière G. and Gouy,M. (1996) WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie, 78, 364–369. [DOI] [PubMed] [Google Scholar]
- 14.Galtier N., Gouy,M. and Gautier,C. (1996) SeaView and Phylo_win: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 12, 543–548. [DOI] [PubMed] [Google Scholar]
- 15.Ponger L. and Mouchiroud,D. (2002) CpGProD: identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics, 18, 631–633. [DOI] [PubMed] [Google Scholar]
- 16.Sharp P.M. and Li,W.H. (1987) The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res., 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulder N.J. Apweiler,R., Attwood,T.K., Bairoch,A., Barrell,D., Bateman,A., Binns,D., Biswas,M., Bradley,P., Bork,P. et al. (2003) The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res., 31, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Etzold T., Ulyanov,A. and Argos,P. (1996) SRS: information retrieval system for molecular biology data banks. Methods Enzymol., 266, 114–128. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 21.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Combet C., Jambon,M., Deléage,G. and Geourjon,C. (2002) Geno3D: automatic comparative molecular modelling of protein. Bioinformatics, 18, 213–214. [DOI] [PubMed] [Google Scholar]
- 23.Lo Conte L., Brenner,S.E., Hubbard,T.J., Chothia,C. and Murzin,A.G. (2002) SCOP database in 2002: refinements accommodate structural genomics. Nucleic Acids Res., 30, 264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pearl M.G., Bennett,C.F., Bray,J.E., Harrison,A.P., Martin,N., Shepherd,A., Sillitoe,I., Thornton,J. and Orengo,C.A. (2003) The CATH database: an extended protein family resource for structural and functional genomics. Nucleic Acids Res., 31, 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westbrook J., Feng,Z., Chen,L., Yang,H. and Berman,H.M. (2003) The Protein Data Bank and structural genomics. Nucleic Acids Res., 31, 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Using CLUSTAL for multiple sequence alignments. Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]
- 27.Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zemla A., Venclovas,C., Fidelis,K. and Rost,B. (1999) A modified definition of SOV, a segment-based measure for protein secondary structure prediction assessment. Proteins, 34, 220–223. [DOI] [PubMed] [Google Scholar]
- 29.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 30.Deléage G., Combet,C., Blanchet,C. and Geourjon,C. (2001) ANTHEPROT: an integrated protein sequence analysis software with client/server capabilities. Comput. Biol. Med., 31, 259–267. [DOI] [PubMed] [Google Scholar]
- 31.Blanchet C., Combet,C., Geourjon,C. and Deléage,G. (2000) MPSA: integrated system for multiple protein sequence analysis with client/server capabilities. Bioinformatics, 16, 286–287. [DOI] [PubMed] [Google Scholar]
- 32.Geourjon C. and Deléage,G. (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci., 11, 681–684. [DOI] [PubMed] [Google Scholar]
- 33.Rost B., Sander,C. and Schneider,R. (1994) Redefining the goals of protein secondary structure prediction. J. Mol. Biol., 235, 13–26. [DOI] [PubMed] [Google Scholar]
- 34.Geourjon C., Combet,C., Blanchet,C. and Deléage,G. (2001) Identification of related proteins with weak sequence identity using secondary structure information. Protein Sci., 10, 788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskowski R.A., McArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst., 26, 283–291. [Google Scholar]
- 36.Thioulouse J., Chessel,D., Dolédec,S. and Olivier,J.M. (1997) ADE-4: a multivariate analysis and graphical display software. Stat. Comput., 7, 75–83. [Google Scholar]
- 37.Perrière G. and Thioulouse,J. (2003) Use of correspondence discriminant analysis to predict the subcellular location of bacterial proteins. Comput. Methods Programs Biomed., 70, 99–105. [DOI] [PubMed] [Google Scholar]
- 38.Combet C., Blanchet,C., Geourjon,C., Deléage,G. (2000) NPS@: Network Protein Sequence Analysis. Trends Biochem. Sci., 25, 147–150. [DOI] [PubMed] [Google Scholar]
- 39.Brunger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,N., Pannu,N.S. et al. (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Cryst., D54, 905–921. [DOI] [PubMed] [Google Scholar]
- 40.Schuler G.D., Epstein,J.A., Ohkawa,H. and Kans,J.A. (1996) Entrez: molecular biology database and retrieval system. Methods Enzymol., 266, 141–162. [DOI] [PubMed] [Google Scholar]