Abstract
Prediction of trans-membrane helices continues to be a difficult task with a few prediction methods clearly taking the lead; none of these is clearly best on all accounts. Recently, we have carefully set up protocols for benchmarking the most relevant aspects of prediction accuracy and have applied it to >30 prediction methods. Here, we present the extension of that analysis to the level of an automatic web server evaluating new methods (http://cubic.bioc.columbia.edu/services/tmh_benchmark/). The most important achievements of the tool are: (i) any new method is compared to the battery of well-established tools; (ii) the battery of measures explored allows spotting strengths in methods that may not be ‘best’ overall. In particular, we report per-residue and per-segment scores for accuracy and the error-rates for confusing membrane helices with globular proteins or signal peptides. An additional feature is that developers can directly investigate any hydrophobicity scale for its potential in predicting membrane helices.
INTRODUCTION
Membrane spanning proteins are vital for cells to function (1,2). However, it is very difficult to experimentally determine high-resolution three-dimensional (3D) structures for these proteins: <50 structures are currently deposited in the Protein Data Bank (PDB) (3,4). C-terminal fusions with indicator proteins (5,6) and from antibody-binding studies (7,8) reveal the location of the helices and the orientation with respect to the protein termini. We refer to these data—slightly incorrectly—as ‘low-resolution structures’. Möller, Apweiler and colleagues at the EBI (9) have carefully hand-selected the results from low-resolution experiments for ∼500 proteins. We have taken the high-resolution set from PDB, added the low-resolution set from the EBI and have filtered the noise from redundancy of very similar proteins by creating the largest possible sub-set that is chosen such that we cannot infer structural similarity between any pair of proteins in that set from sequence alone [sequence-unique subset (10,11)]. Our previous work gives a comprehensive evaluation of transmembrane helix prediction methods based on these sets (10,12). Our automatic benchmarking server accomplishes this by using several different evaluation criteria for evaluating accuracy of the prediction method against proteins of known high- and low-resolution structure. False positives are estimated by applying the method to signal peptides and proteins without trans-membrane helices.
METHODS
Input
The server accepts two types of input from users. Firstly, simple scales reflecting the propensity of residues to form membrane helices, e.g. hydrophobicity scales. Such scales for each amino acid are either uploaded in text format or entered into a form. Secondly, results from novel prediction methods: developers can benefit from the benchmark server by following these steps: (i) download the data sets from our web site (∼2200 proteins, some of which contain membrane helices, some do not); (ii) run your method on all proteins; (iii) upload the predictions to our server in either of two commonly accepted formats. The upload is checked for possible problems that are immediately communicated to the developer. For example, if the predictions contain three-state—helix/non-helix/possible-helix—predictions rather than two state—helix/non-helix—predictions, the user is given a choice to convert all possible-helix residues to non-helix, or convert all possible-helix residues to helix.
Algorithm
When we test hydrophobicity scales, we simply apply the Wimley-White algorithm to turn such scales into predictions of membrane helices (25). New prediction methods (or predictions) are evaluated directly from the data uploaded by the users. A detailed description of the particular scores and schemes explored to measure performance is beyond the scope of this manuscript; they are available in our original publications or on our web site (10,12).
Output
Submissions are tracked through identifiers (IDs) that are shown on all result pages. After the request is queued, the user can either refresh the results page to check the status of the request or follow the link that is emailed to the provided email address when the request is completed. When the results are ready (∼5 min), the user is presented with several tables showing how well the tested method/scale performs in comparison to established methods (Table 1). Results are given separately for (i) high- and (ii) low-resolution membrane proteins, as well as for the discrimination against (iii) globular proteins and against (iv) signal peptides (Fig. 1). In all four resulting tables, several columns show different measures for prediction accuracy and discrimination. Clicking on the column headers will resort the given table by that metric. Clicking on the question mark (?) in the column header names will give a description of the metric. Clicking on the other prediction servers in the row header will give the full name of the server as well as the citation for the source as well as a web link to the server if available. Although the primary format used is the interactive web document described here, the results can also be obtained in non-interactive format. If desired, the results will be emailed in text format along with the link to the interactive results. Additionally, a permanent, non-interactive web document can be generated on the server by clicking a link on the interactive web page. A link to the document is then provided which the user can use to reference the server's results.
Table 1. A list of all advanced methods tested and selected simple hydrophobicity scale methods.
| Method (Reference) | Note |
|---|---|
| DSSP (13) | Program assigning secondary structure |
| HMMTOP (14) | Hidden Markov model predicting transmembrane helices |
| DAS (15) | Optimises use of hydrophobicity plots |
| SOSUI (16) | Uses a combination of hydrophobicity and amphiphilicity preferences to predict membrane helices |
| TMHMM (17) | Embeds a number of statistical preferences and rules into a hidden Markov model |
| PRED-TMR (18) | Uses a standard hydrophobicity analysis with emphasis on detecting the beginnings and ends of helices |
| PHDhtm (19,20) | Combines a neural network using evolutionary information with a dynamic programming optimisation |
| TopPred2 (21) | Averages the GES-scale of hydrophobicity |
| Ben-Tal (22) | Hydrophobicity scale representing free energy of transfer of an amino acid from water into centre of hydrocarbon region of lipid bilayer |
| Kyte-Doolittle (23) | Hydropathy index |
| Wolfenden (24) | Hydration potential |
| Wimley-White (25) | Hydrophobicity scale |
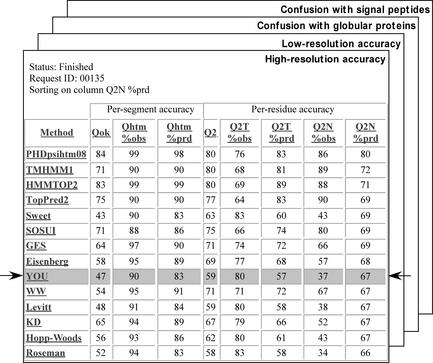
Figure 1.
Sample for server output. Example of one of the four output tables from the server showing where the tested method falls in the ranking of existing methods. Table shown is for accuracy of predicting helices as tested against known high resolution structures. The method named YOU is highlighted in the output and shows the result for the tested method. Hyperlinks re-sort by the given column and also lead to column (scoring metric) and row (prediction method) descriptions.
DISCUSSION
Standard point of reference
The primary goal of this server is to provide users, developers and referees with a standard benchmark evaluation for helical trans-membrane prediction methods in a format that is publicly available and as convenient as possible. The tool may help all not to over-estimate performance and/or to spot strengths and weaknesses of particular methods. The battery of measures for performance that we use encompasses almost all the scores that are found applied in the literature. With the web server, any new algorithm can be tested instantly and seamlessly.
Downside of static benchmark: over-fit to do well on this set only
We might argue that a possible problem with such an easily available and comprehensive method is that someone with enough time on his hands could write a program searching the space of all possible hydrophobicity-like scales in order to optimise the performance on our sets. More generally, developers may over-fit their models. In fact, to some extent, this is a principle problem of any standard data set accepted in the community. However, we challenge that if the scale/method really does consistently better than all methods in respect to all scores, it may indeed capture important aspects of helical membrane proteins. Perhaps more probable is the possibility that one may accidentally over-fit to the benchmark by testing against the benchmark several times during development. To that end, developers can at least reduce the risk of fooling themselves by first testing their final or nearly final method on their own data sets and by then investigating to what extent their results are confirmed by ours.
Ultimate solution: go dynamic
Nevertheless, there is only one way to completely solve the problem, namely test on proteins that could not have been used to develop methods since their experimental structures arrived after the method. This is the concept that we explore through our EVA server evaluating the performance of structure prediction for globular proteins (26–28). However, for globular proteins 10–50 new structures appear in PDB every week. Although this will not become reality for membrane proteins in the foreseeable future, we are currently investigating ways of embedding some dynamic system for the evaluation of membrane predictions into EVA.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Jinfeng Liu and Megan Restuccia (Columbia) for computer assistance; to Chien Peter Chen (Columbia) for his in-depth analysis of membrane helix predictions. Particular thanks to Volker Eyrich for his crucial help with setting up the META-PP and EVA servers without which most of the results presented here would not exist. Thanks to the anonymous reviewer for very detailed, constructive comments. This work was supported by grants RO1-GM63029-01 from the National Institute of Health (NIH) and 1-R01-LM07329-01 from the National Library of Medicine (NLM). Last, but not least, thanks to Amos Bairoch (SIB, Geneva), Rolf Apweiler (EBI, Hinxton), Phil Bourne (San Diego University) and their crews for maintaining excellent databases and to all experimentalists who enabled this tool by making their data publicly available.
REFERENCES
- 1.Thanassi D.G. and Hultgren,S.J. (2000) Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol., 12, 420–430. [DOI] [PubMed] [Google Scholar]
- 2.Truscott K.N. and Pfanner,N. (1999) Import of carrier proteins into mitochondria. Biol. Chem., 380, 1151–1156. [DOI] [PubMed] [Google Scholar]
- 3.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jayasinghe S., Hristova,K. and White,S.H. (2001) MPtopo: A database of membrane protein topology. Protein Sci., 10, 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGovern K., Ehrmann,M. and Beckwith,J. (1991) Decoding signals for membrane proteins using alkaline phosphatase fusions. EMBO J., 10, 2773–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Geest M. and Lolkema,J.S. (2000) Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol. Mol. Biol. Rev., 64, 13–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amstutz P., Forrer,P., Zahnd,C. and Pluckthun,A. (2001) In vitro display technologies: novel developments and applications. Curr. Opin. Biotechnol., 12, 400–405. [DOI] [PubMed] [Google Scholar]
- 8.Traxler B., Boyd,D. and Beckwith,J. (1993) The topological analysis of integral membrane proteins. J. Membr. Biol., 132, 1–11. [DOI] [PubMed] [Google Scholar]
- 9.Möller S., Kriventseva,E.V. and Apweiler,R. (2000) A collection of well characterised integral membrane proteins. Bioinformatics, 16, 1159–1160. [DOI] [PubMed] [Google Scholar]
- 10.Chen C.P., Kernytsky,A. and Rost,B. (2002) Transmembrane helix predictions revisited. Protein Sci., 11, 2774–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rost B. (1999) Twilight zone of protein sequence alignments. Protein Eng., 12, 85–94. [DOI] [PubMed] [Google Scholar]
- 12.Chen C.P. and Rost,B. (2002) Long membrane helices and short loops predicted less accurately. Protein Sci., 11, 2766–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 14.Tusnady G.E. and Simon,I. (1998) Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol., 283, 489–506. [DOI] [PubMed] [Google Scholar]
- 15.Cserzö M., Wallin,E., Simon,I., von Heijne,G. and Elofsson,A. (1997) Prediction of transmembrane α-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng., 10, 673–676. [DOI] [PubMed] [Google Scholar]
- 16.Hirokawa T., Boon-Chieng,S. and Mitaku,S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics, 14, 378–379. [DOI] [PubMed] [Google Scholar]
- 17.Sonnhammer E.L.L., von Heijne,G. and Krogh,A. (1998) In Glasgow,J., Littlejohn,T., Major,F., Lathrop,R., Sankoff,D. and Sensen,C. (eds), Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB98). AAAI Press, Montreal, Canada, Vol. 6, pp. 175–182. [PubMed] [Google Scholar]
- 18.Pasquier C., Promponas,V.J., Palaios,G.A., Hamodrakas,J.S. and Hamodrakas,S.J. (1999) A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng., 12, 381–385. [DOI] [PubMed] [Google Scholar]
- 19.Rost B., Casadio,R., Fariselli,P. and Sander,C. (1995) Prediction of helical transmembrane segments at 95% accuracy. Protein Sci., 4, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rost B., Casadio,R. and Fariselli,P. (1996) Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci., 5, 1704–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman D.M., Steitz,T.A. and Goldman,A. (1986) Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem., 15, 321–353. [DOI] [PubMed] [Google Scholar]
- 22.Kessel A. and Ben-Tal,N. (2002) In Simon,S. and McIntosh,T. (eds), Peptide-Lipid Interactions. Academic Press, San Diego, Vol. 52, pp. 205–253.
- 23.Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- 24.Wolfenden R., Andersson,L., Cullis,P.M. and Southgate,C.C.B. (1981) Affinities of amino acid side chains for solvent water. Biochemistry, 20, 849–855. [DOI] [PubMed] [Google Scholar]
- 25.Jayasinghe S., Hristova,K. and White,S.H. (2001) Energetics, stability, and prediction of transmembrane helices. J. Mol. Biol., 312, 927–934. [DOI] [PubMed] [Google Scholar]
- 26.Eyrich V.A., Marti-Renom,M.A., Przybylski,D., Madhusudhan,M.S., Fiser,A., Pazos,F., Valencia,A., Sali,A. and Rost,B. (2001) EVA: continuous automatic evaluation of protein structure prediction servers. Bioinformatics, 17, 1242–1243. [DOI] [PubMed] [Google Scholar]
- 27.Marti-Renom M.A., Madhusudhan,M.S., Fiser,A., Rost,B. and Sali,A. (2002) Reliability of assessment of protein structure prediction methods. Structure, 10, 435–440. [DOI] [PubMed] [Google Scholar]
- 28.Koh I., Eyrich,V.A., Marti-Renom,M.A., Przybylski,D., Madhusudhan,M.S., Fiser,A., Pazos,F., Valencia,A., Sali,A. and Rost,B. (2003) EVA: evaluation of protein structure prediction servers. Nucleic Acids Res., 31, 3311–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]