Abstract
Multiphoton-targeted photochemistry was used to selectively inactivate the expression of genes in vertebrate cells. A membrane permeable DNA-associating vital dye, ethidium bromide monoacetate (visible wavelength single photon absorption peak at 530 nm) was used to photosensitize chromosomes in dividing cells. A 100-ps infrared laser beam operating at 1.06 microns was focused onto a selected region of a mitotic chromosome corresponding to the sites of the nucleolar (ribosomal) genes. Individual cells followed through mitosis demonstrated a reduction in the number of nucleoli formed in daughter cells that corresponded to the number of nucleolar genes sites irradiated. These results demonstrate the ability to selectively manipulate genes by using the focal point specificity characteristic of multiphoton microscopy. This technique should have wide biotechnology applications both in vitro and in vivo.
Multiphoton (two-photon) scanning laser microscopy has revolutionized the field of functional microscopic biological imaging (1). In particular, the focal point specificity of the technique provides the capability to image objects at the focal plane with little or no excitation at other points in the z axis. At the time that two photon scanning laser microscopy was first described, the authors also suggested that this technique might permit spatially resolved photochemistry. In our early work, we used both single-photon and multiphoton processes to produce subcellular destructive events (2, 3). However, the thermal and shock wave damage produced by the nanosecond-pulsed laser beams often extended beyond the laser focal point, making cell survival and clonability studies difficult. Based on the results of two-photon imaging, we describe multiphoton-targeted photochemistry for selective gene inactivation. This technique provides a complementary functional approach to molecular detection methods such as fluorescence in situ hybridization, such that selectively targeted genetic sites may be inactivated or possibly even activated.
We describe the use of multiphoton-targeted photochemistry to inactivate the ribosomal (nucleolar organizer) genes in cells of the rat kangaroo Potorous tridactylis (PTK2). These cells characteristically remain very flat throughout all stages of mitosis, thus affording clear visualization of the chromosomes, nucleoli, and nuclear envelope (4, 5). The cells were originally derived from a male, having one X-chromosome on which the nucleolar (ribosomal genes) are located in one morphologically recognizable chromosomal location (the secondary constriction). Thus, most of the cells in any cell culture population will have one nucleolus (Fig. 1A). However, approximately 1–5% of the cells in any culture set-up will be near-tetraploid and have two nucleoli per cell (Fig. 1B). Daughter cells of a single nucleolar cell will each have one nucleolus and daughter cells from a double nucleolar cell will have two.
Figure 1.
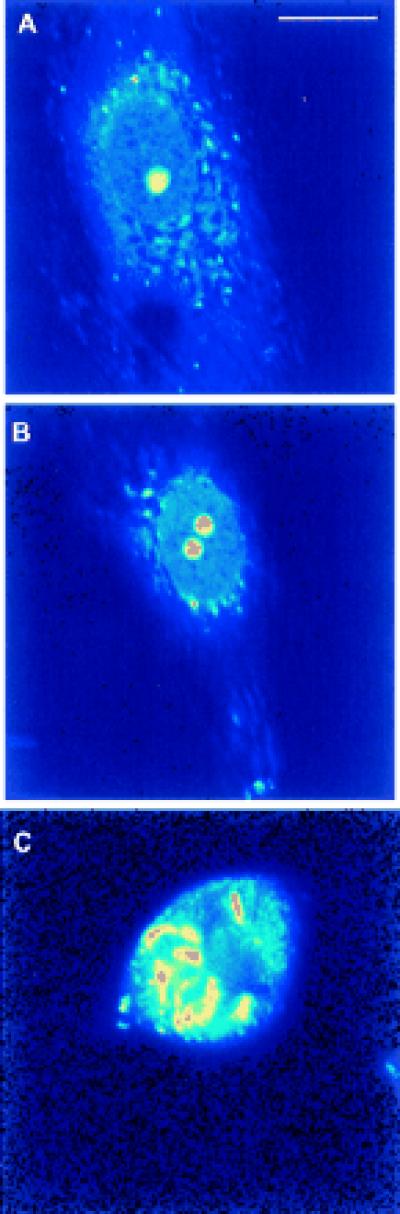
Two-photon fluorescent image of live PTK2 cells vitally stained with ethidium monoazide bromide. (A) An interphase cell with one nucleolus. (B) An interphase cell with two nucleoli. (C) A late prometaphase cell illustrating the high selectivity of the stain for the chromosomes. (Bar = 10 μm.)
In early and mid-prophase, the nucleolus can be seen clearly associated with chromosomes (Figs. 2A and 3A). These chromosomes contain the ribosomal DNA, which is associated with and extends through the nucleolus. Earlier single photon experiments with low power visible and UV lasers, as well as high power multiphoton microplasma-inducing laser beams, demonstrated that irradiation of the nucleolus and the adjacent attached chromosomes results in daughter cells with a corresponding reduction in nucleoli and ribosomal DNA. The reduction in nucleoli was attributable to the inability of the damaged/destroyed nucleolar genes to initiate ribosomal RNA synthesis and subsequently produce a nucleolus at the completion of mitosis (6, 7). However, in these early studies, the single photon thermal and multiphoton microplasma-induced shock wave damage extended outside the laser focal point, producing secondary damage that reduced cell survival. In this report, we demonstrate the use of multiphoton-targeted photochemistry to inactivate a genetic site in a confined spatial region with subsequent 100% cell survival.
Figure 2.
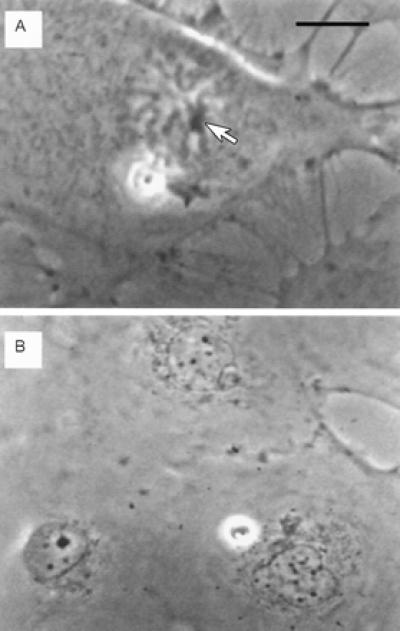
Live phase contrast images of pre- and post-irradiation single nucleolar cell treated with ethidium monoazide bromide. (A) Pre-irradiation mid-prophase cell. One phase dense nucleolus can be seen clearly associated with condensed chromosomes (arrow). The nucleolus plus the closely associated chromosomes were exposed to the laser irradiation to ensure exposure of all of the ribosomal DNA. (Bar = 10 μm.) (B) The two daughter cells 24 h after laser exposure. Neither daughter cell has a nucleolus. Both have small micronuclear bodies that are typically found scattered throughout the nucleus. Note the size of the nucleolus and the three small nuclear bodies in the adjacent normal cell in this field. In earlier experiments (see ref. 9), inactivation of all of the ribosomal DNA by single-photon methods produced daughter cells with small nuclear bodies identical to the ones observed here. (Bar in A = 15 μm in B.)
Figure 3.
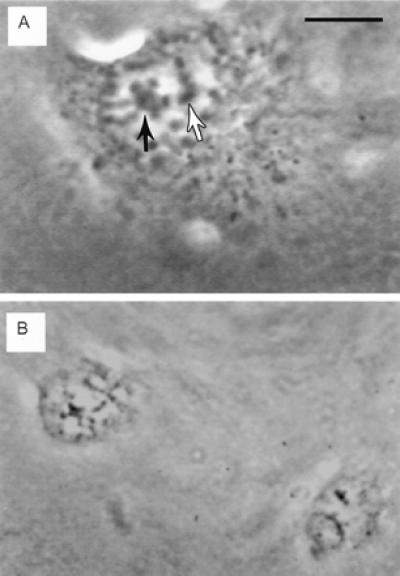
Live phase contrast images of pre- and post-irradiation double nucleolar cell treated with ethidium monoazide bromide. (A) Prophase cell with partially condensed chromosomes associated with two nucleoli (arrows). This cell was followed from the initiation of prophase to track the two nucleoli during the process of chromosome condensation. One nucleolus and its associated chromosome regions were irradiated (black arrow). (Bar = 10 μm.) (B) Two daughter cells 8 h after completion of mitosis. Both cells have only one nucleolus. The partially condensed chromatin visible in both nuclei generally disappears by 12 h after mitosis, when the cell enters the S phase of the cell cycle. (Bar in A = 15 μm in B.)
Methods
PTK2 (Potorous tridactylis) cells were grown in standard tissue culture flasks and then were seeded into Rose tissue culture chambers for experimentation using well established techniques (4, 5). Digital video mages were stored for each experimental and control cell before laser exposure and for up to 24 h after mitosis to facilitate precise identification of cells. Cells were photosensitized by using ethidium monoazide bromide (phenanthridium: 3-amino-8-azido-5-ethyl-6-phenyl,bromide) obtained from Molecular Probes. The single photon absorption spectrum of the ethidium monoazide exhibits a broad single maximum between 475 and 530 nm. This dye intercalates into the DNA of living cells and, after exposure to visible photons (488 nm), undergoes a single-photon photochemical-induced covalent bond to the DNA (8). We theorized that spatially confined two-photon induced photochemical inactivation of genes should be possible using this dye in combination with the appropriate infrared wavelength.
The dye was obtained in powder form, was solubilized in 50 mM potassium phosphate buffer at pH 7, and was introduced in standard cell culture medium at a concentration of 10 μg/ml of culture medium. Cells were incubated at 37°C, 10% CO2 in the dye-culture medium for 12 h before experimentation. Cell culture chambers were wrapped in aluminum tin foil until placed on the microscope stage to protect them from ambient light.
The laser microscope constructed specifically for these gene inactivation studies used a pulsed (100 ps) Coherent Antares Nd-Yag laser operating at 70 MHZ (Coherent Radiation, Palo Alto, CA). The 1.06-μm wavelength beam was introduced into a Zeiss Axiovert inverted microscope through the standard epifluorescent port from which all of the collection optics had been removed. Before entry into the microscope, the beam passed through an optical beam expander so that the diameter of the laser fully filled the rear aperture of the microscope objective. A dichroic filter (>90% reflectance, 700 nm to 1.2 microns; > 70% transmittance, 500–650 nm) was mounted in the epifluorescent slider positioned between the microscope objective and the rest of the microscope optical system. The 1.06-μm excitation beam entered the rear aperture of either a ×100 or ×63 numerical aperture 1.4 Zeiss phase contrast objective. The beam was focused to a near-diffraction spot of 1-μm diameter at a specific target region inside a single cell. Power measurements were made on the laser beam before entry into the microscope as well as exiting the microscope objective by using a Coherent LaserMate model 33-0191 power meter.
Whole cell two-photon images were made by using a laboratory-built two photon scanning microscope. The system consisted of a 5-W solid-state, frequency-doubled Nd:YVO4 (Verdi, Coherent) pumped titanium:sapphire (Ti:Al2O3) laser (Mira, Coherent) used as the two-photon excitation source. The Ti:Al2O3 laser was tuned to 800 nm, allowing two-photon imaging. The mode-locked, 100-fs, 76-MHz pulse train exiting the Ti:Al2O3 laser was attenuated by neutral density filters that enabled output power levels to stay below tissue damage thresholds by reducing the average power at the sample (typically 5–10 mW). The laser beam was scanned across a sample placed on a microscope (Zeiss Axiovert 100) using an X-Y scanner (Series 603X, Cambridge Technology, Cambridge, MA), which was controlled by a digital card developed at the Laboratory for Fluorescence Dynamics at the University of Illinois at Urbana-Champaign. The two-photon fluorescence was detected by using a single-photon counting detection system, which consisted of a photomultiplier tube (R5600P, Hamamatsu, Middlesex, NJ). The scan size was 256 × 256 pixels, with a typical pixel size of 0.15 μm.
Results and Discussion
The two-photon excitation capability of the picosecond laser microscope was readily demonstrated in PTK2 cells treated with the ethidium monoazide bromide vital dye. The 1.06-μm wavelength laser beam was focused onto individual chromosomes of dividing live cells. Bright red-orange fluorescence was detected visually (Fig. 4 A and B) and was subsequently collected over 5-s periods by using an Acton Research (Acton, MA) SpectraPro-150 spectrometer coupled to a cooled CCD detector (Princeton Instruments TE/CCD-576E) mounted on the laser microscope. A log/log plot (slope = 1.95) of fluorescence intensity versus laser power was derived by stepwise attenuation of the laser beam before entry into the microscope (Fig. 5).
Figure 4.
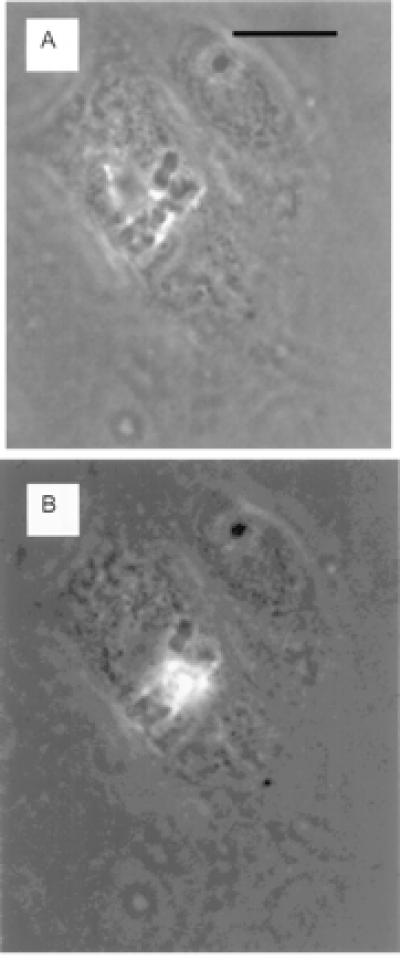
(A) A mitotic cell with chromosomes clearly visible aligned at the metaphase plate. (B) The same cell with the chromosomes in the center of the metaphase plate exhibiting two-photon fluorescence during exposure to the 1.06-μm-wavelength laser beam focused through the ×63 microscope objective. Cells were exposed to ethidium monoazide bromide at 10 μm/1 ml for 24 h. (Bar = 15 microns.)
Figure 5.
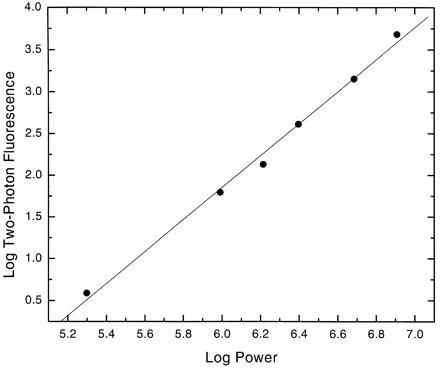
Two-photon log-log plot fluorescence (slope = 1.95) of ethidium monoazide bromide bound to DNA in live mitotic Protorous tidactylis cells. Average laser powers entering the microscope on the abscissa were 100 mW to 1.1 W with excitation wavelength = 1.06 μm. Fluorescence emission was 520–700 nm.
After demonstration that the picosecond laser microscope could induce two photon events, nine single nucleolar and three double nucleolar PTK2 cells treated with ethidium monoazide bromide were irradiated. Power density in the 1-μm focal spot was in the range of 1.02–1.27 × 1011 W/M2. This is equivalent to 7.18 × 1021 photons/M2 for a 100-ps pulse and 3.27 × 1031 photons/M2 for 60 s of exposure. Each cell had its target region exposed to the laser beam for 30–60 s. Cell positioning under the laser beam was controlled by moving a digital-controlled 0.2-μm resolution microscope stage (Ludl Electronic Products, Hawthorne, NY) while viewing the image of the target cell on a television monitor. The nucleolus and associated chromosomes were exposed to the focused infrared laser beam by scanning the target zone under the laser beam during the 30-to 60-s exposure period.
In single nucleolar cells, the nucleolus and its attached chromosomes were irradiated. In double nucleolar cells, only one nucleolus and its attached chromosomes was irradiated. Each individual cell was followed through mitosis for up to 24 h so that the nucleolar content of the daughter cells could be assayed. Nineteen control single nucleolar cells without exposure to ethidium monoazide bromide were irradiated under identical conditions to the experimental cells.
All 12 of the experimental cells proceeded through mitosis and produced daughter cells with reduced nucleolar number. The nine single nucleolar prophase cells produced daughter cells without any normal nucleolus (Fig. 2). The cells had small dark nuclear bodies that are typical of cells whose ribosomal DNA have been inactivated (6, 7, 9). The double-nucleolar prophase cells having one nucleolus and associated chromosomes irradiated produced daughter cells with one nucleolus each (Fig. 3). All 19 control cells produced daughter cells with one nucleolus.
Conclusions
The experimental results demonstrate that multiphoton-targeted photochemistry can be used to alter a selected genetic region in living cells with 100% cell survival. We have used a well studied cytogenetic system to validate our hypothesis. This approach could be applied to other gene targets in which there may be either a morphological chromosome marker or a sequence-specific molecular probe conjugated to a photon-absorbing molecule. This technique should have wide application in basic studies on gene regulation, functional genomics, and targeted gene therapy.
Acknowledgments
Two photon imaging microscopy was conducted in the National Institutes of Health Biotechnology LAMMP Resource at the University of California at Irvine. The two photon inactivation experiments were conducted on the picosecond microscope system built by the lead author (M.W.B.) while on sabbatical in the Department of Bioengineering, University of California at San Diego. Professor Shu Chien is gratefully acknowledged for the space provided for these studies. The Office of Naval Research, the Department of Energy, and the National Institutes of Health Bioengineering Initiative provided for additional support for this research.
References
- 1.Denk W, Strickler J H, Webb W W. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 2.Berns M W. Biophys J. 1976;16:973–977. doi: 10.1016/S0006-3495(76)85747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calmettes P P, Berns M W. Proc Natl Acad Sci USA. 1983;80:7197–7199. doi: 10.1073/pnas.80.23.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berns M W, Aist J, Edwards J, Strahs K, Girton J, McNeill P, Rattner J B, Kitzes M, Hammer-Wilson M, Liaw L H, et al. Science. 1981;213:505–513. doi: 10.1126/science.7017933. [DOI] [PubMed] [Google Scholar]
- 5.McNeill P A, Berns M W. J Cell Biol. 1981;88:543–553. doi: 10.1083/jcb.88.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns M W, Chong L K, Hammer-Wilson M, Miller K, Siemens A. Chromosoma. 1979;73:1–8. doi: 10.1007/BF00294839. [DOI] [PubMed] [Google Scholar]
- 7.Berns M W, Cheng W K. Exp Cell Res. 1971;69:185–192. doi: 10.1016/0014-4827(71)90324-7. [DOI] [PubMed] [Google Scholar]
- 8.Bolton P H, Kearns D R. Nucleic Acids Res. 1978;5:4891–4903. doi: 10.1093/nar/5.12.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berns M W, Floyd A D, Adkisson K, Cheng W K, Moore L, Hoover G, Ustick K, Burgott S, Osial T. Exp Cell Res. 1972;75:424–432. doi: 10.1016/0014-4827(72)90449-1. [DOI] [PubMed] [Google Scholar]


