Abstract
The GeneSeqer@PlantGDB Web server (http://www.plantgdb.org/cgi-bin/GeneSeqer.cgi) provides a gene structure prediction tool tailored for applications to plant genomic sequences. Predictions are based on spliced alignment with source-native ESTs and full-length cDNAs or non-native probes derived from putative homologous genes. The tool is illustrated with applications to refinement of current gene structure annotation and de novo annotation of draft genomic sequences. The service should facilitate expert annotation as a community effort by providing convenient access to all public plant sequences via the PlantGDB database, a simple four-step protocol for spliced alignment and visually appealing displays of the predicted gene structures in addition to detailed sequence alignments.
INTRODUCTION
Since the inception of the USA National Plant Genome Initiative (NPGI) in 1998 and similar programs in other countries, the infrastructure for plant genetic, molecular and applied research has dramatically changed. Traditional, ‘one gene at a time’ approaches are now routinely complemented by whole genome analyses that evaluate the features, properties and functions of particular genes or gene products in the context of the entire genome and proteome, as well as the larger evolutionary context. Sequence data have been accumulating from three major sources: whole genome sequencing and assembly, nearly completed for Arabidopsis thaliana (1) and two cultivars of rice (2,3); genome survey sequencing, in progress for complex genomes such as maize (4); and expressed sequence tags (ESTs), currently exceeding two million entries in dbEST (http://www.ncbi.nlm.nih.gov/dbEST/). In view of the recommended goals for the NPGI in the next 5 years (5), this data flow will likely continue, with a focus on complete sequencing of ‘reference species’ (Arabidopsis, rice, maize, Medicago truncatula and tomato), draft sequencing of other selected species and further EST and full-length cDNA sequencing.
These sequencing efforts are the prelude to the goal of complete structural and functional characterization of the plant transcriptome. Accurate annotation of the genomic sequences has remained a bottleneck in the pursuit of this goal. Even for A.thaliana, more than 2 years after publication of most of the genome, genome annotation is still incomplete and in need of refinement (6,7). There are at least two major causes of this problem. First, the pace of sequencing requires initial automated gene structure annotation, and such ab initio annotation has limited accuracy (8). Secondly, refined annotation using cDNA or EST evidence (9–12) is restricted by incomplete sampling of transcripts.
We have set up the GeneSeqer@PlantGDB web server to help address the plant genome annotation bottleneck. The server integrates the up-to-date public plant sequence records available at our database PlantGDB (http://www.plantgdb.org) with the unique spliced alignment capabilities of the GeneSeqer application (13), allowing for the robust annotation of plant genomic sequences using both native and homologous ESTs, cDNAs and protein sequences. The server is meant to complement offline annotation pipelines by providing convenient access to tools and data for experts on particular genes or gene families to annotate thoroughly those genes. For Arabidopsis, the server is already linked to a prototype community annotation database that should facilitate propagation of such expert annotation.
DESCRIPTION OF WEB SERVICE
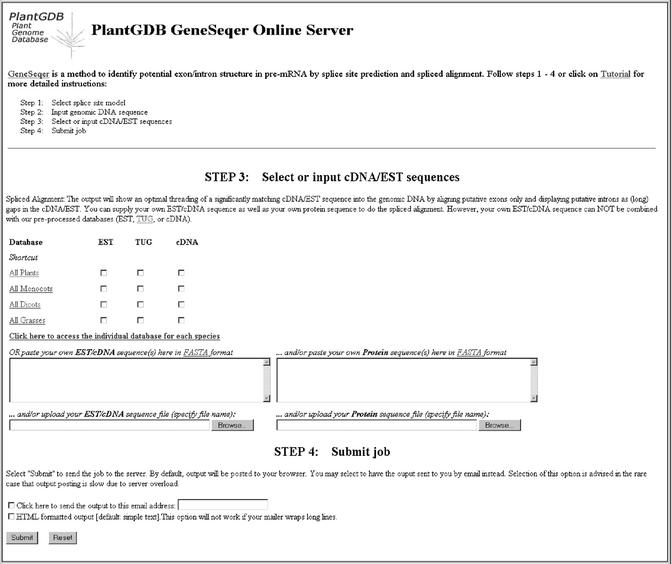
The GeneSeqer@PlantGDB server relies on a simple four-step protocol (Fig. 1). Initially, the user selects an appropriate model for splice site prediction (currently limited to Arabidopsis as a representative dicot and maize as a representative monocot) and supplies the genomic DNA sequence. The sequence can be uploaded from a local disk, pasted or retrieved by a GenBank accession number. The third step is selection or input of the probes for spliced alignment. The user may choose the current collections of ESTs, EST assemblies (tentative unique gene fragments or TUGs) or full-length cDNAs. The collections may be chosen to encompass all available plant sequences, broad taxonomic groups or specific species only. In addition, users may supply their own probes, which may also include potential protein homologs of a suspected gene product encoded in the input genomic sequence.
Figure 1.
GeneSeqer@PlantGDB. The server implements a simple four-step protocol. Steps 1 and 2 were omitted from the figure for clarity.
Upon submission, the server returns interactively to the browser or via email results of the spliced alignment. Because the GeneSeqer algorithm involves complete dynamic programming alignment for promising probes, very large applications would not be practical over the web. Therefore, at most 500 alignments are returned per genomic input sequence. The web output of the GeneSeqer@PlantGDB server includes graphic summaries of the predicted gene structures (including alternative transcripts) and encoded proteins. The protein sequences are linked to the NCBI Blast Server (14) to facilitate queries to the protein database for potential homologs, which in turn may be used as probes for further spliced alignment (for example, to extend partial EST evidence).
APPLICATIONS
We discuss two typical applications. The first application is refined annotation of a previously reported gene structure based on additional spliced alignment evidence. The second example illustrates iterative annotation of a novel genomic sequence.
Example 1: refined analysis of existing annotation
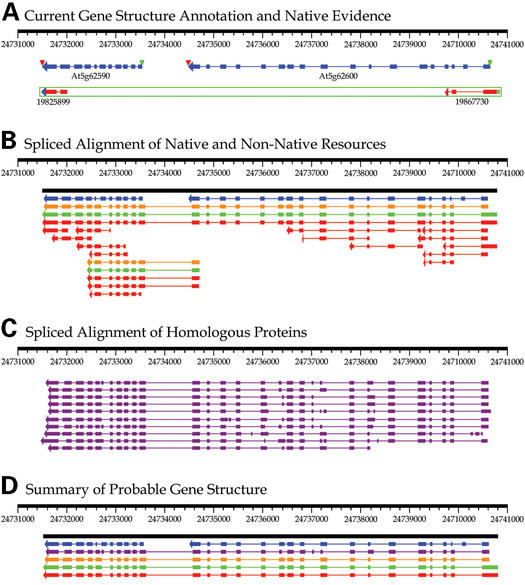
Static gene structure annotation, such as that established for the Arabidopsis genome sequence, is inherently unreliable with respect to updated sequence collections. In particular, matching ESTs, cDNAs or proteins precisely supporting a given gene structure may not have been available at the time of annotation. Thus, annotation of such a gene region based mainly on ab initio gene prediction is likely to be imprecise. This problem, referred to as annotation lag, is demonstrated in Figure 2 for a region of chromosome five of the A.thaliana genome. Figure 2A shows two gene models for this region (At5g62590, annotated as an unknown protein, and At5g62600, a putative protein with similarity to transportin-SR) and a single pair of Arabidopsis ESTs spanning the entire region. The EST mapping is available at AtGDB (http://www.plantgdb.org/AtGDB; 9), as is a specialized GeneSeqer server (http://www.plantgdb.org/cgi-bin/AtGeneSeqer.cgi) that directly uploads the Arabidopsis genomic sequence of interest (but is otherwise equivalent to GeneSeqer@PlantGDB).
Figure 2.
Refined analysis of an existing gene annotation. (A, B and C) depict three stages in the annotation of gene structure for a region of A.thaliana chromosome five. In each panel, spliced alignments and inferred (established) gene structures are represented by arrows extending from the first exon to the last, pointing in the most probable direction of transcription. Exons are represented as boxes connected by introns shown as single lines. (A) In this display, which is available for all A.thaliana genome regions at http://www.plantgdb.org/AtGDB/, spliced alignments originating from native (Arabidopsis) ESTs are shown in red. Following the AtGDB convention, ESTs are identified by their GenBank gi number, 5′-ESTs are marked by a green dot, 3′-ESTs are marked by a blue arrow and clone pair ESTs are bounded by a green box. In this example, there are only two matching ESTs, representing the 5′- and 3′-sequences of RIKEN clone RAFL07-12-J11. The current gene annotations, as established by AGI (Arabidopsis Genome Initiative), are shown in blue, with start and stop codons labeled with green and red triangles, respectively. (B) This graphic, generated using the GeneSeqer@PlantGDB web service, summarizes the results of spliced alignment using the PlantGDB ‘All Plants’ EST and cDNA collections. Spliced alignments of ESTs and cDNAs, depicted in red, can be attributed to the following sources: cDNA: 1 Glycine max; ESTs: 6 Medicago truncatula, 5 Glycine max, 2 A.thaliana, 1 Lotus japonicus, 1 Sorghum bicolor, 1 Triticum aestivm. Alternative gene structures are shown in green (representing consistent predictions from multiple ESTs) and long open reading frames in the predicted gene structures are indicated in orange. Established gene annotation, as reported in GenBank, is shown in blue. (C) All gene structures shown are the results of GeneSeqer@PlantGDB spliced alignments using putative homologous proteins of non-Arabidopsis origin. (D) summarizes the most probable gene structure prediction for this region. The blue, orange, green and red structures represent respectively the established gene annotation, the longest predicted open reading frame, the predicted gene structure and the consensus transcribed sequence spliced alignment (derived from B). The purple structure represents the alignment with the most closely related protein, a Drosophila protein (GenBank gi: 20177035) with high similarity to vertebrate transportin-SR proteins.
The spliced alignment of only two native ESTs provides minimal evidence as to the overall gene structure in this region, although confirming coding potential. In addition, the ESTs provide evidence that the currently proposed gene annotation for the region is invalid, because both ESTs are denoted as members of the same cDNA clone (RAFL07-12-J11; EST gi:19867730 representing the 5′-sequence and EST gi:19825899 representing the 3′-sequence). Thus, there is most likely a single gene spanning this region as opposed to the two predicted gene annotations. Analysis of a species' transcriptome using only native EST resources may preclude the annotation of less prevalently expressed genes. Similarly, genes with differential expression across certain conditions or tissue types may lack EST evidence or even remain undiscovered unless EST sampling is very deep. This example illustrates, however, how inclusion of homologous ESTs and proteins of non-native origin enhance the coverage attainable by spliced alignment and can improve the accuracy of gene annotation.
Figure 2B shows the results of spliced alignment of the ‘All Plant’ EST and cDNA sets maintained at PlantGDB. Fifteen non-Arabidopsis sequences could be reliably aligned. The tiling of the spliced alignments covering the entire region supports the existence of a single gene in the region. Strikingly, an open reading frame spans all 27 predicted exons. The inferred protein sequence of this open reading frame was used as a query to the NCBI non-redundant protein database via the BLAST link of the GeneSeqer Web output. From the BLAST results, the top 10 putative homologous proteins (from species other than Arabidopsis, including Drosophila melanogaster, zebra fish, mouse and human) were selected from alignments with expectation values <1×10−10. Spliced alignment of these protein sequences, as shown in Figure 2C, was achieved through subsequent GeneSeqer@PlantGDB analysis (for detailed procedure see http://www.plantgdb.org/AtGDB/tutorial/GSQ_ATGDB.php). Figure 2D summarizes the evidence, strongly supporting this region to encompass a single gene for a transportin-SR protein.
Example 2: annotation of a large genomic sequence with limited native resources
Genomic sequence assemblies submitted by high-throughput sequencing projects as draft quality sequences are a great resource for the community even prior to (complete) annotation. For example, a researcher may identify a BAC clone by hybridization with a probe representing a gene of interest. Rather than waiting for annotation of the corresponding sequence by the sequence providers, this researcher will welcome tools to annotate the region encoding his or her gene. Of course, such annotation may still be difficult for the same reasons confounding gene structure annotation on a large scale. However, spliced alignment via GeneSeqer@PlantGDB may provide the answers in particular cases. To illustrate applications of this type, we briefly discuss annotation of a segment of genomic sequence from Sorghum bicolor (GenBank accession AF503433).
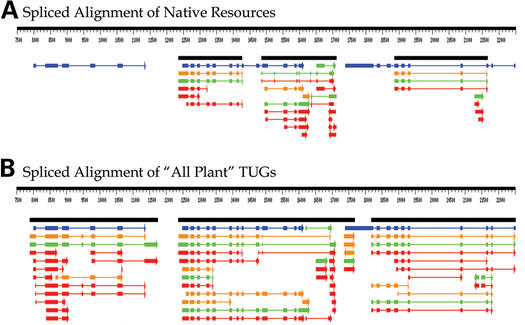
Given the large size (142 kb) of the Sorghum sequence in this example, while using ‘All Plant’ ESTs and cDNAs for spliced alignment as in the previous example is feasible, a more efficient approach is the use of the ‘All Plant’ TUG option. The TUG collection consists of consensus sequences representing the clustering and assembly of each species' EST and cDNA sets (see http://www.plantgdb.org/ESTCluster/progress.html for details), thus removing redundancy of EST representations. (After initial annotation using the spliced alignment of the TUG consensus sequences, a more detailed analysis of individual regions may always be performed to visualize constituent spliced alignments or to probe for interesting biological properties such as evidence of alternative splicing.) In this example, five new gene models were identified. Figure 3 shows gene structure prediction in a representative 15-kb segment. The three loci putatively represent a mitochondrial carrier (tricarboxylate transport) protein, subunit 1 of a cleavage stimulation factor and a serine threonine kinase based on Blastp analysis against the NCBI non-redundant protein database. For a more detailed description of this example and analysis results from the entire BAC, see http://www.plantgdb.org/tutorial/.
Figure 3.
Large-scale annotation by spliced alignment. (A and B) show the output of GeneSeqer@PlantGDB for segment 7500 to 22500 of the S.bicolor bacterial artificial chromosome (BAC) with GenBank accession number AF503433. Graphical representations are as in Figure 2. (A) shows the results of GeneSeqer@PlantGDB analysis using only the Sorghum EST and cDNA collections. (B) displays results of analysis from the identical region now using the ‘All Plants’ TUGs collection.
SUMMARY
The GeneSeqer@PlantGDB web service provides a convenient workbench interface to the tools necessary for complex gene structure annotation tasks. Immediate access to expansive transcribed sequence data collections and interactive visualization of spliced alignment results gives users the ability to generate high-quality gene structure annotation without specific bioinformatics training or experience. At the user's discretion, high-quality annotations may be shared with the research community through services for third party annotation such as the GenBank/EMBL/DDBJ TPA division (15–17) and the User Contributed Annotation (UCA) system of AtGDB (see http://www.plantgdb.org/AtGDB). The Web service is not meant to replace offline annotation pipelines. All constituent programs are freely available to the academic community (http://bioinformatics.iastate.edu/bioinformatics2go/), including a more elaborate tool for visualizing gene prediction evidence from both ab initio and spliced alignment algorithms (MyGV, 18).
Acknowledgments
ACKNOWLEDGEMENT
This work was supported in part by NSF Plant Genome Research Projects grants DBI-0110254 and DBI-9872657.
REFERENCES
- 1.The Arabidopsis Genome Initiative. (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 2.Goff S.A., Ricke,D., Lan,T.H., Presting,G., Wang,R., Dunn,M., Glazebrook,J., Sessions,A., Oeller,P., Varma,H. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92–100. [DOI] [PubMed] [Google Scholar]
- 3.Yu J., Hu,S., Wang,J., Wong,G.K., Li,S., Liu,B., Deng,Y., Dai,L., Zhou,Y., Zhang,X. et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79–92. [DOI] [PubMed] [Google Scholar]
- 4.Chandler V.L. and Brendel,V. (2002) Update: maize genome sequencing project. Plant Physiol., 130, 1594–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The national plant genomics initiative: objectives for 2003–2008. Plant Physiol., 130, 1741–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haas B.J., Volfovsky,N., Town,C.D., Troukhan,M., Alexandrov,N., Feldman,K.A., Flavell,R.B., White,O. and Salzberg,S.L. (2002) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol., 3, research0029.1–0029.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brendel V. and Zhu,W. (2002) Computational modeling of gene structure in Arabidopsis thaliana. Plant Mol. Biol., 48, 49–58. [PubMed] [Google Scholar]
- 8.Pavy N., Rombauts,S., Déhais,P., Matheé,C., Ramana,D.V., Leroy,P. and Rouzé,P. (1999) Evaluation of gene prediction software using a genomic data set: application to Arabidopsis thaliana sequences. Bioinformatics, 15, 887–899. [DOI] [PubMed] [Google Scholar]
- 9.Zhu W., Schlueter,S.D. and Brendel,V. (2003) Refined annotation of the Arabidopsis thaliana genome by complete EST mapping. Plant Physiol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kan Z., Rouchka,E.C., Gish,W.R. and States,D.J. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res., 11, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheelan S.J., Church,D.M. and Ostell,J.M. (2001) Spidey: a tool for mRNA-to-genomic alignments. Genome Res., 11, 1952–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gemünd C., Ramu,C., Altenberg-Greulich,B. and Gibson,T.J. (2001) Gene2EST: a BLAST2 server for searching expressed sequence tag (EST) databases with eukaryotic gene-sized queries. Nucleic Acids Res., 29, 1272–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Usuka J., Zhu,W. and Brendel,V. (2000) Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics, 16, 203–211. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J. and Wheeler,D.L. (2003) GenBank. Nucleic Acids Res., 31, 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoesser G., Baker,W., Van Den Broek,A., Garcia-Pastor,M., Kanz,C., Kulikova,T., Leinonen,R., Lin,Q., Lombard,V., Lopez,R. et al. (2003) The EMBL Nucleotide Sequence Database: major new developments. Nucleic Acids Res., 31, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazaki S., Sugawara,H., Gojobori,T. and Tateno,Y. (2003) DNA Data Bank of Japan (DDBJ) in XML. Nucleic Acids Res., 31, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu W. and Brendel,V. (2002) Gene structure identification with MyGV using cDNA evidence and protein homologs to improve ab initio predictions. Bioinformatics, 18, 761–762. [DOI] [PubMed] [Google Scholar]