Abstract
Information about the secondary and tertiary structure of a protein sequence can greatly assist biologists in the generation and testing of hypotheses, as well as design of experiments. The PROTINFO server enables users to submit a protein sequence and request a prediction of the three-dimensional (tertiary) structure based on comparative modeling, fold generation and de novo methods developed by the authors. In addition, users can submit NMR chemical shift data and request protein secondary structure assignment that is based on using neural networks to combine the chemical shifts with secondary structure predictions. The server is available at http://protinfo.compbio.washington.edu.
BACKGROUND
Protein structure mediates protein function in biological processes that are essential for the survival and development of an organism. We have developed a web server, PROTINFO (http://protinfo.compbio.washington.edu), which predicts the tertiary and secondary structure of a protein, given its amino acid sequence. There are three categories of methods for three-dimensional (tertiary structure) modeling: comparative modeling (CM), fold recognition (FR) and de novo prediction (AB). In the comparative modeling and fold recognition categories, the methodologies rely on the presence of one or more evolutionarily related template protein structures that are used to construct a model; the evolutionary relationship can be deduced from sequence similarity (1–4) or by ‘threading’ a sequence against a library of structures and selecting the best match (5–7). For both approaches, a sequence alignment between the target protein to be modeled and the evolutionarily related protein with known structure is used to create the initial or seed model. In the de novo category, there is no strong dependence on database information and prediction methods are based on general principles that govern protein structure and energetics (8–12). The categories vary in difficulty and consequently methods in each of these categories produce models with different levels of accuracy relative to the experimental structures.
METHODS USED IN THE PROTINFO SERVER
The three-dimensional modeling methods use software developed as part of the RAMP suite of programs and are based on our published research (11,13–18). The source code for the RAMP software, along with more detailed documentation, is accessible from our software distribution server (http://software.compbio.washington.edu).
Comparative modeling using RAMP
The quick method carries out a sequence-only search using a variety of methods and then uses the ‘hits’ returned as seeds for a multiple sequence alignment. Initial models are then built for each alignment to a template and the resulting models are scored using an all-atom function (14,19). Loops and side chains are built on the best scoring models using a frozen approximation (15). The slow method (not available publicly at present) does a sophisticated graph-theory search to mix and match between various main chain and side chain conformations (14).
During the searches, templates with ≥95% sequence identity to the submitted sequence are usually ignored [because this could represent the same structure in the Protein Data Bank (PDB) (20)]. To generate a model where the alignment between the submitted and template sequences has sequence identity ≥95%, one may submit the alignment and template structures explicitly.
This approach is likely to produce the best models when the relationship between the submitted and template proteins is clearly discernible (≥30% sequence identity). Even though models are built if the sequence identity is lower, they are likely to contain errors (see ‘Performance and caveats’ section below).
Fold recognition and generation using RAMP
The RAMP software uses a de novo method for generating a particular fold for any given sequence. To make this work successfully, the correct fold has first to be recognized. For now, the comparative modeling methods described above are used to identify tenuous sequence relationships (hopefully representing distant homologues). Alternatively, one can submit a template structure as a potential fold (for example, based on the function). Once a template has been chosen, it is then used as a ‘beacon’ to guide the simulations and a large number of structures are generated that resemble the template. From this ensemble, the best-scoring structure is selected. The latter part of the procedure is similar to the de novo approach described below.
The structural alignment between the final structure generated for the submitted sequence and the template is used as another alignment choice for traditional comparative modeling. Thus, each sequence is modeled in two ways. The advantage of the de novo fold generation approach is that it does away with the issue of alignment and loop building.
This approach is likely to be most useful when the relationship between a submitted sequence and its corresponding template protein is not perceptible by sequence comparison methods (≤20% sequence identity) and where the sequence is limited to a single domain of <200 residues. Although models will be built if the sequence identity is much higher, they are not likely to be as good as the models built using the comparative modeling approach described above.
De novo prediction using ramp
If there are no related templates to a submitted sequence and/or if the sequence has the appropriate length (∼100 residues), then it may be modeled using a de novo protocol (17). Our general paradigm for predicting structure involves sampling the conformational space such that native-like conformations are observed and then selecting them using a hierarchical filtering technique with many different scoring functions. We explore combinations of different representations/move sets with a combined Monte Carlo/genetic algorithm search method for exploring protein conformational space and combinations of a variety of all-atom scoring function ‘filters’ to identify biologically relevant conformations (17). This approach is likely to be most useful for small sequences.
Secondary structure assignment using PsiCSI
This method uses neural networks to translate NMR chemical shifts into secondary structure information [somewhat similar to CSI (21)] and combines it with sequence-based predictions [akin to Psipred (22)]. It has a sustained three-state average accuracy of 89% on a rigorously jackknifed test set of 92 proteins for which NMR chemical shift information was publicly available (18).
INPUT AND OUTPUT FORMATS AND BEHAVIOR
Input formats and behavior
Sequences must be specified in a single line using the one-letter amino acid notation. Splitting up longer sequences into domains if knowledge of the domain boundaries is available is prudent. This is because the complexities of most calculations are generally exponentially proportional to the lengths of the sequences and most prediction methods are calibrated to work on domains. The programs currently perform a limited amount of automatic domain parsing, which will be enhanced in the future. Very short (≤30 residues) and very long sequences are not likely to generate reliable predictions. Any PDB files submitted optionally must generally start with residue 1 and the residues must be numbered consecutively without any chain breaks. There is some support for cleaning up the PDB files submitted. Chemical shifts for PsiCSI secondary structure predictions are submitted in the form of NMR-STAR files.
Output formats and behavior
Following the convention used in the experiments on the Critical Assessment of Structure Prediction methods (CASP), up to five models for each three-dimensional prediction method (CM, FR, DN) may be returned (in CASP format). Because some of the components of comparative modeling and fold recognition methods are the same, it makes sense to request that both methods be applied to a sequence instead of submitting two separate requests. Under certain conditions (no clear relationship to a template is discerned, for example), both methods may be executed by the PROTINFO server regardless of the method requested.
Detailed output is available both as part of the file that is emailed back to the recipient, as well as the directories in which the data were generated. The output of the secondary structure prediction using PsiCSI consists of the sequence, the secondary structure assignment at that position and the overall confidence of the assignment. Finally, to facilitate interpretation of more ambiguous assignments, the program also outputs the relative propensities for each of the three secondary structure states at each position.
PERFORMANCE AND CAVEATS
Performance at CASP5/CAFASP3
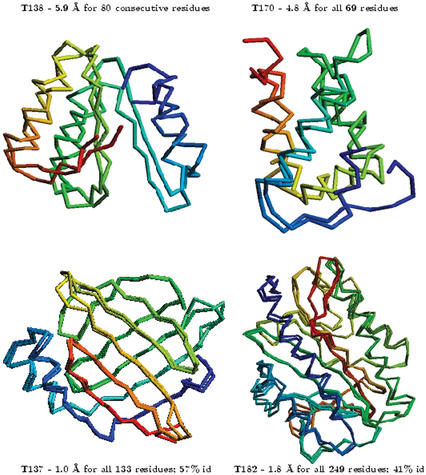
Protein structure prediction methods are rigorously evaluated by the Critical Assessment of Structure Prediction (CASP, and CAFASP for ‘fully automated’) experiments held every 2 years (23) (http://predictioncenter.llnl.gov). This was motivated in part by claims in the literature of the protein-folding problem being ‘solved’ without producing tangible benefits, because most of the ‘solutions’ included a strong dependence on the test set. These experiments evaluate prediction techniques by asking modelers to construct models for a number of protein sequences before the experimental result is known, over a period of 3–4 months. We have taken part in all five CASP experiments, including the most recent one (CASP5), which finished in December 2002 (11,17,24,25). The results provide a benchmark as to what level of model accuracy we can expect from our methodologies, which are among the most competitive at these experiments. Figure 1 gives some examples of our CASP predictions. At CASP5, we made predictions for all 67 targets using our automated server (an experimental answer was made available to us for 35 of these proteins). For the 14 (out of 17) comparative modeling targets that had sequence identities ranging from 50 to 20% to the template used, we produced models ranging from 1.0 to 6.0 Å RMSD (between the Cα atoms of the model and the corresponding experimental structure) for all or large parts of the protein, with model accuracies scaling fairly linearly with respect to percentage sequence identity (that is, the higher the sequence identity, the better the prediction). We made de novo predictions for 25/35 targets. For proteins with lengths >100 residues, predictions were made only for 100–150 residues after identifying putative domains. In all but three cases, we produced models with accuracies ranging from 4.0 to 6.0 Å Cα RMSD for 60–100 residue fragments (or the entire protein). The results represent a slight improvement in most cases compared to our CASP4 results, which show a similar trend and are described in further detail elsewhere (17). The primary difference is that these predictions were made in a completely automated fashion and there is greater consistency in the results, particularly in the de novo predictions for targets that are defined as ‘new fold’ by the CASP assessors.
Figure 1.
Examples of selected CASP5 de novo (top) and comparative modeling (bottom) predictions made by the PROTINFO server. The superposition of the model and the corresponding experimental structure is shown, along with the Cα RMSD relative to the experimental structure. The percentage identity of the alignment between the target and template sequences is given for the bottom two comparative modelling targets. Models as accurate as 1.0 Å Cα RMSD are produced for easy comparative modelling targets. For small proteins or domains without detectable sequence homology to known structures, our de novo methods consistently produce topologically accurate models 4.0–6.0 Å Cα RMSD for 60–100 residue fragments (or the entire protein).
Caveats and analysis of the PROTINFO server
Based on the preliminary analysis of the CASP5/CAFASP3 results, the following observations can be made about the use of this server. We recommend using the PROTINFO servers with these guidelines in mind.
Both the comparative modeling and fold generation methods rely on the selection of an appropriate template (and an alignment for the former). The current default methods implemented for doing this seem to work best only on the easiest (clearly recognizable homology) cases. They usually fail on intermediate and the most difficult cases. To get around this problem, we recommend first submitting the sequence to the Bioinfo MetaServer (http://bioinfo.pl) and obtaining the best result (alignment as well as template) from the 3D Jury method. The 3D Jury is a meta-predictor that combines the results of the other predictors and makes a consensus evaluation, using a flexible interface that lets a user define the choice of individual servers to use, as well as the method used to pick the consensus model. This template choice (with all the main chain atoms present) as well as the alignment (for comparative modeling) can be submitted as input to the PROTINFO server.
Even if the template is correctly specified, the fold generation method is not likely to work better for larger proteins, or when the template and the submitted target proteins are highly similar, relative to the traditional comparative modelingapproach.
The de novo method does indeed seem to consistently produce topologically accurate structures for small proteins (≤100 residues) and/or fragments of a protein, even for the most difficult cases. For the one case (t138; Fig. 1) where chemical shift data were available, the three-state secondary structure accuracy using PsiCSI was 13% higher (resulting in a 52% error reduction) than using sequence-only methods. This undoubtedly helped our de novo prediction. Because of the way the models are generated, our ‘MODEL 1’ might not necessarily correspond to the best structure. There are filters that can be used to figure out which is the best model (including the SCORE record). The model numbering does not always correlate with its assigned score.
Calculation times and current usage
The secondary structure predictions made using PsiCSI only take ∼2–3 min on a single processor (in fact, PsiCSI can be used semi-interactively) and are returned rapidly. Although only the ‘quick’ versions of the tertiary structure prediction methods are publicly available, they are usually executed on a cluster with hundreds of processors for each sequence modeled. Each submitted sequence through the PROTINFO server will, however, be limited to a single processor (and there are only a limited number of processors made available for public use). Our goal is to ensure that the prediction time for each sequence is <24 h (comparative modeling predictions will most likely take only a few hours), but this depends on how many people submit sequences. Because the computation time required is much longer for longer sequences, the software is executed on different processors for different length ranges (<200, 200–400, >400 residues). Thus people submitting long sequences may have to wait much longer on average before their sequence is pulled for processing, compared to people submitting short sequences. There is a feature to monitor the progress of submissions.
Currently, the server receives 10–20 sequences per day, which is far greater than the computational resources allocated to handling them (at least for the three-dimensional modeling portion). Thus a response might not be sent for several days. Nonetheless, given the detailed model-building capabilities of the server, we feel it a useful resource for the study of protein structure. We expect to dedicate more computational resources in the near future.
Future work
Enhancements planned for the near future include a module to predict the tertiary structure of proteins given noisy or limited NMR data along with our de novo methods, a module to assess binding energies/affinities of substrate–protein interactions and a module for protein–protein docking calculations.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Patrice Koehl for his efficient FORTRAN code used for constructing protein structures and calculating RMSDs, and Michael Levitt for providing us with the ENCAD energy minimization software. We also thank past and present members of the Levitt, Moult and Samudrala groups for their intellectual guidance and support that helped in the development of this software. This work was supported in part by a Searle Scholar Award and NSF Grant DBI-0217241 to R.S. and the University of Washington's Advanced Technology Initiative in Infectious Diseases.
REFERENCES
- 1.Doolittle R. (1981) Similar amino acid sequences: chance or common ancestry? Science, 214, 149–159. [DOI] [PubMed] [Google Scholar]
- 2.Greer J. (1990) Comparative modeling methods: application to the family of the mammalian serine proteases. Proteins, 7, 317–334. [DOI] [PubMed] [Google Scholar]
- 3.Sander C. and Schneider,R. (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 4.Murzin A. and Bateman,A. (1997) Distant homology recognition using structural classification of proteins. Proteins, 29S, 105–112. [DOI] [PubMed] [Google Scholar]
- 5.Bowie J., Lüthy,R. and Eisenberg,D. (1991) Method to identify protein sequences that fold into a known three-dimensional structure. Science, 253, 164–170. [DOI] [PubMed] [Google Scholar]
- 6.Jones D., Taylor,W. and Thornton,J. (1992) A new approach to protein fold recognition. Nature, 258, 86–89. [DOI] [PubMed] [Google Scholar]
- 7.Flöckner H., Domingues,F. and Sippl,M. (1997) Protein folds from pair interactions: a blind test in fold recognition. Proteins, S1, 129–133. [DOI] [PubMed] [Google Scholar]
- 8.Lee J., Liwo,A., Ripoll,D., Pillardy,J. and Scheraga,J. (1999) Calculation of protein conformation by global optimization of a potential energy function. Proteins, S3, 204–208. [DOI] [PubMed] [Google Scholar]
- 9.Ortiz A., Kolinkski,A., Rotkiewicz,P., Ilkowski,B. and Skolnick,J. (1999) Ab initio folding of proteins using restraints derived from evolutionary information. Proteins, S3, 177–185. [DOI] [PubMed] [Google Scholar]
- 10.Osguthorpe D. (1999) Improved ab initio predictions with a simplified flexible geometry model. Proteins, S3, 186–193. [DOI] [PubMed] [Google Scholar]
- 11.Samudrala R., Xia,Y., Huang,E. and Levitt,M. (1999) Ab initio protein structure prediction using a combined hierarchical approach. Proteins, S3, 194–198. [DOI] [PubMed] [Google Scholar]
- 12.Simons K., Bonneau,R., Ruczinski,I. and Baker,D. (1999) Ab initio structure prediction of CASP3 targets using ROSETTA. Proteins, S3, 171–176. [DOI] [PubMed] [Google Scholar]
- 13.Samudrala R., Xia,Y., Levitt,M. and Huang,E. (1999) A combined approach for ab initio construction of low resolution protein tertiary structures from sequence. In Altman,R., Dunker,A., Hunter,L., Klein,T. and Lauderdale,K. (eds), Proceedings of the Pacific Symposium on Biocomputing. World Scientific Press, pp. 505–516. [DOI] [PubMed] [Google Scholar]
- 14.Samudrala R. and Moult,J. (1998) A graph-theoretic algorithm for comparative modeling of protein structure. J. Mol. Biol., 279, 287–302. [DOI] [PubMed] [Google Scholar]
- 15.Samudrala R. and Moult,J. (1998) Determinants of side chain conformational preferences in protein structures. Protein Eng., 11, 991–997. [DOI] [PubMed] [Google Scholar]
- 16.Xia Y., Huang,E., Levitt,M. and Samudrala,R. (2000) Ab initio construction of protein tertiary structures using a hierarchical approach. J. Mol. Biol., 300, 171–185. [DOI] [PubMed] [Google Scholar]
- 17.Samudrala R. and Levitt,M. (2002) A comprehensive analysis of 40 blind protein structure predictions. BMC Struct. Biol., 2, 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong-Hung L. and Samudrala,R. (2003) Accurate and automated assignment of secondary structure with PsiCSI. Protein Sci., 12, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samudrala R. and Moult,J. (1998) An all-atom distance dependent conditional probability discriminatory function for protein structure prediction. J. Mol. Biol., 275, 895–916. [DOI] [PubMed] [Google Scholar]
- 20.Berman H., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T., Weissig,H., Shindyalov,I. and Bourne,P. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart D., Sykes,B. and Richards,F. (1992) The chemical shift index: a fast and simple method for the assignment of protein secondary structure through nmr spectroscopy. Biochemistry, 31, 1647–1651. [DOI] [PubMed] [Google Scholar]
- 22.Jones D. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol., 292, 195–202. [DOI] [PubMed] [Google Scholar]
- 23.Moult J., Hubbard,T., Fidelis,K. and Pedersen,J. (1999) Critical Assessment of Methods of Protein Structure Prediction (CASP): Round III. Proteins, S3, 2–6. [PubMed] [Google Scholar]
- 24.Samudrala R., Pedersen,J., Zhou,H., Luo,R., Fidelis,K. and Moult,J. (1995) Confronting the problem of interconnected structural changes in the comparative modeling of proteins. Proteins, 23, 327–336. [DOI] [PubMed] [Google Scholar]
- 25.Samudrala R. and Moult,J. (1997) Handling context-sensitivity in protein structures using graph theory: bona fide prediction. Proteins, 29S, 43–49. [DOI] [PubMed] [Google Scholar]