Abstract
Sequence annotation is essential for genomics-based research. Investigators of a specific genomic region who have developed abundant local discoveries such as genes and genetic markers, or have collected annotations from multiple resources, can be overwhelmed by the difficulty in creating local annotation and the complexity of integrating all the annotations. Presenting such integrated data in a form suitable for data mining and high-throughput experimental design is even more daunting. DNannotator, a web application, was designed to perform batch annotation on a sizeable genomic region. It takes annotation source data, such as SNPs, genes, primers, and so on, prepared by the end-user and/or a specified target of genomic DNA, and performs de novo annotation. DNannotator can also robustly migrate existing annotations in GenBank format from one sequence to another. Annotation results are provided in GenBank format and in tab-delimited text, which can be imported and managed in a database or spreadsheet and combined with existing annotation as desired. Graphic viewers, such as Genome Browser or Artemis, can display the annotation results. Reference data (reports on the process) facilitating the user's evaluation of annotation quality are optionally provided. DNannotator can be accessed at http://sky.bsd.uchicago.edu/DNannotator.htm.
INTRODUCTION
Today, more than 95% of the human genome has been sequenced. To organize and understand the biological meanings of the sequence data, annotation is required. Celera (http://www.celera.com/), Map Viewer from NCBI (National Center for Biotechnology Information) (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/map_search), Genome Browser (http://genome.ucsc.edu/) from UCSC (University of California Santa Cruz) and Ensembl from the Sanger Center (http://www.ensembl.org/) provide a variety of annotations on the genomic sequence assembled by NCBI or elsewhere (Celera). The word ‘annotation’ is used here (throughout the paper and the informatics tools to be described) to mean mapping of varieties of features to genomic DNA sequences. The annotated features from these public or private resources are very valuable as the basis of experimental design and data analysis, especially for projects searching for susceptibility genes of genetic diseases. However, existing annotation may not be sufficient for particular gene-hunting projects, and investigators, for several reasons, will often have to do some annotation in their own laboratories.
Firstly, an important factor demanding local annotation activities in investigative laboratories is related to the quality of current genomic sequence assembly, as well as quality and quantity of annotation source data. For the genomic sequence assembly, individual laboratories focusing on certain chromosome regions could have a better regional assembly than that in the public database. The assembly quality issue would persist for a while, especially for ‘difficult’ regions like 15q. Study of so-called ‘genomic disorders,’ which arise from structural rearrangements of chromosomes, represents a more extreme case, requiring the investigator to re-organize the regional sequence to mimic that involved in those disorders. Public annotation would generally not address such case-specific regional annotation problems as successfully as laboratories focused on these problems. It is expected that major biological findings, such as novel genes, SNPs and regulatory elements, will be discovered in individual laboratories. As new experimental technologies, especially the high-throughput techniques, are developed and improved, it is expected that individual laboratories would have enormous amounts of data and local discoveries as original annotation building blocks to be mapped into regional genomic sequences. Even for the data in the public domain, individual laboratories are more flexible and devoted to collecting data from different resources for a specific genomic region. Most importantly, in view of the potential delay in periodic updates of the public annotation, laboratories focusing on a genomic region might have more urgent needs of regional annotation than the general research community. Recently, several papers (1–3) presented the needs for genome-wide re-annotation as more knowledge and better annotation technologies were developed after the original annotation. This rationale also holds true for regional annotation.
Secondly, integrating of annotation from different resources or migrating it from one sequence to an updated one would be required to provide researchers with the updated sequence along with complete annotation data.
Thirdly, there are gaps between public annotation efforts and the interests of the individual researcher. Public annotation efforts may not be interested in certain annotation that researchers need. For example, sequence-related laboratory data, such as primers, oligos, amplicons and so on, can be important for the management of research resources and progress of a project, but many of these data would not be of general interest and therefore would not likely be annotated by public efforts.
Putting all this together, we see an increasing need for batch custom annotation on genomic regions, by which the investigator annotates his/her own source to his/her own target sequence. Several tools are available to do annotation over a user's genomic region sequence. Genotator (4), NIX (Williams et al., http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix/), GeneMachine (5), GAIA (6), Alfresco (7), GESTALT (8), RUMMAGE (9) and Oak Ridge National Laboratory Genome Analysis Pipeline (http://compbio.ornl.gov/tools/pipeline) provide integrated annotation, including mapping of known or predicted genes and/or regulatory elements by running multiple gene-prediction programs and searching against static public databases. However, few of them incorporate methods for SNP mapping, which is essential for positional cloning projects for complex diseases. None of these systems takes source data supplied by the end user, unless the user can modify the databases in the annotation system.
Some individual programs can be used for annotation of certain types of custom source data over the user's own gDNA sequence. For example, BLAST (10) is good at homolog sequence searching. Sim4 (11), est_genome (12) and Spidey (13) were designed for cDNA–gDNA (genomic DNA) alignment to define the intron–exon structure of a gene. e-PCR (14) can be used to map STSs. However, most of these programs provide unique output formats, which cannot be directly converted into standard format annotation. Freeware, such as Artemis (15), Sequin (16) and some commercial software like Vector NTI, provide a good interface to do manual annotation. However, as currently provided, they are not a solution for batch annotation.
Distributed Annotation System (DAS) (17) is one of the most advanced systems for annotation data management and exchange, but it does not provide an automatic annotation method. Genome Browser, in which the user can supply a ‘custom track,’ requires creation of input data for this custom function, which is more challenging. Genome Browser provides the BLAT function (18) so that an end user can roughly map custom sequences onto the public genome sequence, but only the public assembly can be used as the annotation target. Nonetheless, BLAT alone is not sufficient for batch annotation because no filter or selection is provided, so it would require additional data processing for sequences with multiple hits or partial matches in the genome, and for obtaining accurate positions of SNPs, since a BLAT alignment shows a sequence range, and not a single nucleotide position. BLAT also would not work for mapping very short oligos because 21 bp is its minimum search limit. Therefore, the problem of batch annotation of the user's source data over preferred gDNA sequence remains.
The existing software packages described attempt to provide solutions for de novo annotation rather than preserving existing annotation. In laboratories enormous amounts of time could be wasted on re-annotating an updated sequence, and the manual annotation currently performed is particularly error-prone as well as time-consuming.
Besides the problem of local batch custom annotation, most existing tools do not provide outputs in a standard format suitable for data archiving or further data mining. Consequently, it would require additional work to do data mining or large-scale experiment design based on the annotation results from these systems.
To aid the investigator who has abundant locally organized source data in spreadsheet, database or FASTA format files, and a need to perform regional batch annotation for data management or efficient experimental design, DNannotator takes in user-supplied source data and target sequences and produces annotation output in standard formats that can be viewed and analysed in other programs and database systems. The user who already has annotation in GenBank format on an old version sequence but wants to use the up-to-date gDNA sequence or who needs to integrate varieties of annotations into one common sequence platform can use DNannotator's annotation migration function.
Linkage studies (19–23) present evidence of bipolar disorder and schizophrenia susceptibility gene(s) at 13q. We performed annotation of this region as a case example for DNannotator. STSs, selected SNPs and primers are annotated into different versions of gDNA assemblies of this sizeable genomic region.
METHODS
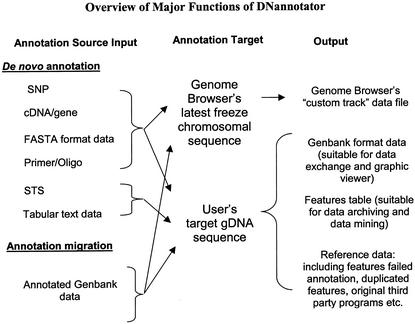
DNannotator is a web application running under the Apache HTTP server in a UNIX environment. Web pages provide an interface for uploading data files and setting analysis parameters. A set of Perl/CGI scripts behind the web pages is used for most of the data processing, including formatting data, calling external programs, such as BLAST, BLAT, Sim4 and e-PCR, and sending results to the user by email. The central components of DNannotator are functions performing annotations using the approaches described below. Some other associated Perl, Java and C programs were used to provide accessory functions, such as manipulating data format, extracting feature-related sequences and cleaning up data files. The workflow of DNannotator's major annotation functions is illustrated in Figure 1. Currently, DNannotator annotates SNPs, primers, gene exons, STSs and other user-specified source data onto user-provided or Genome Browser's latest freeze gDNA sequence.
Figure 1.
Overview of major functions of DNannotator. Using input source data of STSs, SNPs, primers, general FASTA or tab-delimited text data, de novo annotation can be carried out on target gDNA sequences through different function modules of DNannotator. A single GenBank formal data file can be used as input to perform annotation migration.
Input data
Annotation source data
Because spreadsheets and databases are often used in laboratories to archive annotation source data, including sequence and related information for all the features (or annotation elements), DNannotator takes a tab-delimited text file, which can be easily exported from a spreadsheet or a database, as input for annotation of SNPs and primers. Since gene cDNA sequences are normally stored in FASTA format, DNannotator was designed to take FASTA format input for gene exon mapping. With the original annotation provided in GenBank format, DNannotator can perform annotation migration based on sequence identities to create annotation of another related sequence.
Target gDNA sequence
DNannotator accepts FASTA format gDNA sequence as an annotation target. If annotation to Genome Browser's latest freeze is performed, DNannotator supplies the chromosomal gDNA sequence.
Output data
All major annotation functions of DNannotator produce a set of optional outputs, including annotation results in GenBank format data and in tab-delimited text format, a list of feature elements that failed annotation, a list of features mapped to multiple locations, and annotation basis or evidence such as original and parsed-and-filtered BLAST, BLAT results or Sim4 original results, and so on. The tab-delimited text features table includes gDNA sequence ID, feature name, feature type (exon, variation, etc.), physical start and end positions, and the orientation of the feature. To facilitate quality control, the user can also elect to obtain additional annotation quality-related information, such as the BLAST match identity percentage value of alignment for each annotation feature. One special output, a custom track data file for Genome Browser's map, can be created for those annotation functions targeting Genome Browser's latest freeze chromosomal sequence. This data file can be directly loaded into Genome Browser and viewed side by side with other annotations supplied by Genome Browser.
De novo annotation
DNannotator uses two basic approaches to carry out de novo annotation: (a) BLAST-match-based approach (SNP and primer annotations are implemented in this fashion), (b) parsing of outputs of third-party software (gene exon annotation is implemented by parsing Sim4 or BLAT results; STS is annotated with e-PCR).
BLAST-match-based annotation
In a BLAST-match-based (BMB) annotation procedure, BLAST or BLAT searches all the feature sequences against the target gDNA sequence. The BLAST format result is automatically parsed and double-filtered using a user-adjustable threshold. The first filter is a combination of match size and percentage of identity and size difference between query sequence and matching fragment. The second filter can find only the best match out of all the matches that survived the first filter. Both filters can be adjusted to accommodate the needs of detecting sequence homologs. Based on the matches that survive the two filters, physical positions in the target gDNA of all features from the source data are calculated taking into account all the gaps introduced in each BLAST alignment block. Descriptions of features in source data are formatted and mapped onto target sequences following these computations. Partial matches of source target sequence alignments or those crossing several non-continuous alignment blocks are not used for annotation. Therefore, some highly polymorphic STSs and genes cannot be annotated by the BMB method.
DNannotator implements four different entry points for annotation using this BMB approach for the sake of the different source data content (Fig. 1). Two of them are specialized for annotation of SNPs' or primers' source data in formatted tab-delimited text. SNP source data require SNP ID, polymorphic allele and flanking sequences. Primer source data require primer name and primer sequence. Besides these functions, DNannotator can also take source sequence data with the associated information, including feature name, feature type, and orientation, as input, in either formatted tab-delimited text or FASTA format. Ideally all other features could be annotated in these two fashions.
DNannotator provides both BLAT and BLAST as choices for BMB annotation.
Annotation by parsing of outputs of third-party software
A e-PCR analysis (modified to produce orientation information of primer pairs) is used to do STS mapping using data regarding STS primers' sequence and amplicons' size range, which can either be obtained from NCBI's dbSTS (supplied by DNannotator) or specified by users. Parsing the e-PCR output, uniform STS annotation results are created.
DNannotator provides two methods for exon analysis: Sim4-based annotation with parsing of the Sim4 results and BLAT-based annotation with parsing of its PSL format output from BLAT.
Annotation migration between sequences
DNannotator uses a combination of BMB and e-PCR-based approaches to migrate annotation from one annotated sequence to another related one. With GenBank format of the original, annotated sequence as input, information and corresponding sequence fragments are extracted, excluding the ‘repeat_region’ features. Then, de novo annotation on the new target sequence is performed. Most features are annotated in BMB fashion, because most of the accurate annotations in the source data should be built on continuous gDNA sequences. For very short (<40 bp) features such as primer/oligo or small exons, an additional flanking sequence (40 bp) is automatically attached to the feature sequence so that high stringency BMB annotation can be performed without losing the short alignments. With the difficulty of annotating certain features by BMB as mentioned above and difficulty in querying very long sequences in BLAST or BLAT, features like ‘gene’, ‘STS’ and all features longer than 10 kb each are annotated in e-PCR fashion by searching a 25 bp sequence at both ends of the feature regions. Concerned about false-positive annotation from querying such short sequences, we tested longer end sequences (30 and 40 bp), but they resulted in a higher rate of false-negative annotation because of sequence differences located in the primer region, which was the key factor for e-PCR-based annotation (data not shown). In a number of annotation migrations performed, we did not observe any false-positive annotation using 25 bp end sequences.
Annotation to the latest freeze of the Genome Browser map
Considering that many users will not construct their own regional gDNA sequence, DNannotator provides annotation to the latest freeze of Genome Browser's chromosomal sequence. The functions are similar to those of fully customized annotation, which requires a gDNA sequence from users as discussed above.
RESULTS
Linkage evidence strongly implies the existence of susceptibility genes for bipolar disorder and schizophrenia in 13q32-33. Annotation of this region was performed for a research project on bipolar disorder (19–21) and for evaluation of annotation quality of DNannotator.
Case study: annotation of genomic DNA sequence of 13q32-33
Target gDNA sequence
Sequences covering, but not limited to, the region between D13S122 and D13S779 from NCBI's assemblies build 30 (NT_009952.10, called NCBI30, 25 Mb), our manually assembled sequence of the region (named TA, 17 Mb, unpublished), and Genome Browser's November 2002 freeze chromosome 13 were used as annotation targets.
De novo annotation
In all, 49 cDNA sequences (including four genes discovered or extended locally), 548 primers used in our lab, 1600 SNPs selected from public databases, 157 SNPs or insertions/deletions identified in our lab and STSs from NCBI's UniSTS were mapped onto the above three different versions of gDNA assemblies of this region or chromosome. All 49 transcripts were recognized and annotated as about 560 exons in each. Five extremely short exons (<10 bp) considered unreliable calls from Sim4 were labeled with warning messages. A few transcripts have a more than 10 bp cDNA sequence not covered by the exon report. In the case of SNP mapping, ∼1750 SNPs were mapped successfully to all assemblies. Only one false-positive mapping, rs2009772, was reported as duplicated when the second filter was turned off. However, once turned on, no false-positive mapping was produced. Fewer than seven SNPs failed the annotation and were reported by DNannotator. Five of them show false-negative results because of excessive gaps introduced in the BLAST matches, which broke up the BLAST alignment block and failed the annotation. The others were a result of a low quality gDNA sequence over a small region. More than 400 STSs were annotated by DNannotator and some duplicated annotations were observed. Six of them (D13S158, D13S174, D13S278, D13S281, D13S286, D13S128) were caused by redundant records in the UniSTS database. Two distinct markers mapped to different places (1.6 kb away) on 13q are both named D13S128. Since UniSTS data has been pre-computed in the Genome Browser map, we did not provide STS mapping for it. In the case of mapping primers, 12 out of 548 primers were found to be part of repeat sequence, especially in Alu, by the DNannotator utility ‘screen primer for repeats.’ Four of the 12 primers have more than 100 identical copies in NCBI30. These 12 primers in repeats were excluded from mapping. Fewer than 40 primers could not be mapped into assemblies because most (32) of them were designed for amplification on cDNA, BAC clone vectors, genes on other chromosomes or were modified by adding extra sequence tails. The rest of the failures are located at a region containing either low-quality or polymorphic sequences. Detailed analysis of annotation results can be obtained in the supplementary material.
Annotation migration
Annotation migration was performed to transfer local annotation from TA to NCBI30. After using DNannotator's utility to merge all annotations with sequence data to create one GenBank format data file, all 3268 features annotated in TA created by DNannotator including 1751 SNPs, 513 primer, 556 exons and 448 STSs, were migrated into NCBI30 in one shot by the annotation migration function. Of the 3268 features to be migrated, only three STSs, two primers and one exon failed to be transferred and they were reported by DNannotator. All the failures were caused by sequence quality or polymorphism below the filter threshold.
Annotation results
All annotation results in GenBank format can be viewed graphically by Artemis (Fig. 2) and other viewers accepting GenBank format data such as Vector NTI. Annotations for Genome Browser's chromosomal sequence can be viewed in Genome Browser (Fig. 3). Feature tables in tab-delimited text can be directly imported into an MS-Access database or MS-Excel spreadsheet for data archiving and data mining.
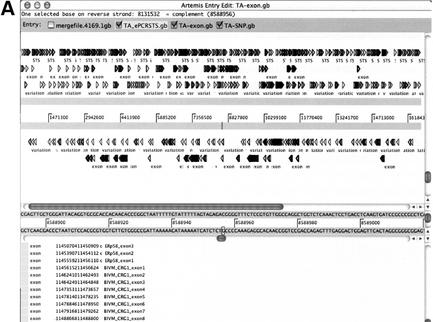
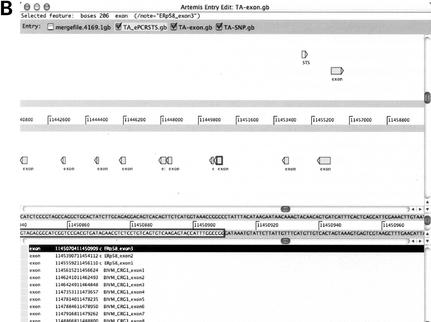
Figure 2.
Graphic presentation of annotated TA assembly. Individual annotated gb-header files (GenBank format data without sequence body) produced by DNannotator were directly read in as ‘Entry’ by Artemis. Exon, STS and SNPs were displayed (A) Broad view of annotations in TA assembly. (B) Enlarged view of annotations. The window is composed of three panels. Features listed in the lower panel, graphic icons in the upper panel and sequence data in the middle panel are internally linked to each other. Clicking one will highlight corresponding elements in the other panels.
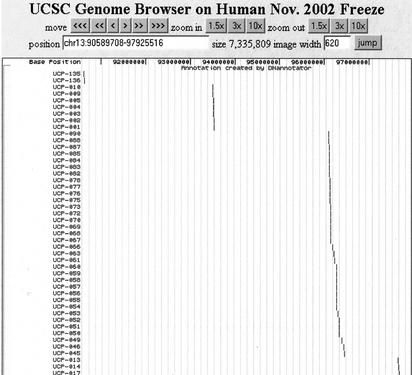
Figure 3.
Viewing annotation created by DNannotator in Genome Browser. The SNPs discovered locally were mapped to Genome Brower's latest freeze of chromosome 13 by DNannotator, with an output of the custom track data file of Genome Browser, which can be directly viewed side by side with all other annotations provided by Genome Browser.
CONCLUSIONS AND DISCUSSION
DNannotator uses two approaches, BMB and third-party programs, to annotate user-supplied source data onto user-customized gDNA sequences or Genome Browser's chromosomal sequences. It gives the user much flexibility to make batch de novo annotation on interesting regions. DNannotator also provides a robust solution for migrating annotation from one sequence to another. In a ∼20 Mb region of chromosome 13, more than 3000 elements, including primers, SNPs and STSs were annotated in three different gDNA assemblies and migrated between assemblies. The annotation results demonstrated that this system was generally robust and accurate with only a small number (<1%) of failures using the default setting of DNannotator. The user supplies only source data and/or the target sequence and an email address.
Through analysis of the individual failures, it was observed that sequence quality and regional polymorphism density are the major factors affecting annotation quality. Sequence errors or polymorphisms in either source data or target gDNA sequence at the places where features are located could lower identity of the sequence match, which is the key for both BMB and e-PCR-based annotation. Sequence quality is especially important for annotation of primers and STSs, because these two annotations rely on very short sequence matches, while high stringency conditions have to be applied to overcome the problem of excessive hits in the genome for short sequences.
DNannotator does have some limitations: it might not be able to carry out de novo annotation of features with very long sequences because BLAST has difficulty querying them. We recommend using the BMB approach to map features shorter than 10 kb. Fortunately, most features we are dealing with daily are short elements such as SNPs, exons and primers. Some limitations of DNannotator are related to the inherent limitations of other third-party programs. The e-PCR-based method could suffer from the same problem of mapping primers. STSs such as D13S259 from UniSTS could not be mapped in any of the assemblies by the e-PCR-based method, because mismatches could be found in the primer binding sequence. Sim4 tends to make false annotation for small exons or skips some very short exons and creates low identity exons. As only a small genomic region is analysed as the target, DNannotator would not detect features that might have multiple copies in the whole genome. In other words, DNannotator can report duplicated features in regional annotation but would not guarantee the uniqueness throughout the genome for those features not reported for duplication.
To minimize labor costs while maintaining the highest accuracy of annotation, DNannotator supplies a series of warning messages and annotation reference data for the user's review. Most annotation functions provide separate outputs for failed and duplicated features in the annotation process. Most annotation raw data are provided as an optional reference output. Therefore, the user can easily skim through large amounts of annotation results to find the very limited number (1–5% from our observation) of potentially unreliable results and correct them manually or re-annotate them in DNannotator with adjusted settings. We consider this a major advantage of DNannotator over some existing public annotations, which only present the final annotation results making evaluation of annotation quality difficult. We found that a number of SNPs and STSs in public databases are not mapped by public annotation, but, unfortunately, no additional information is available. Without comparison, we could not know what was missed in the public annotation.
DNannotator provides functions for transferring annotation from one sequence to another, alleviating the problem of preserving annotation of updated sequences. Because it is based on the individual features that are the basic building blocks of annotation, annotation migration relies on the quality of local sequences harboring those individual features, rather than the quality of assembly of the whole sequence. Thus, annotation migration could be successful for most features even if the assembly structure is significantly changed.
Annotation migration is preferred over de novo annotation when a variety of features are already incorporated in a GenBank format data file because the user does not need to format source data nor carry out de novo annotation using different modules of DNannotator for different feature types. We observed that most of the features of one sequence could be transferred faithfully to another by annotation migration.
Many choices of format are available for annotation data presentation. For example, NCBI GenBank provides GenBank, ASN.1, Graphics and XML formats. Of this list GenBank format is the only compact format without complicated tags and therefore can be viewed by a simple text editor, which is more accessible to biologists. At the same time, a number of graphic viewers like Artemis are available for viewing GenBank format data. Hence, DNannotator uses standard GenBank format as a common platform for both input and output sequence data. Moreover, with the tab-delimited text and GenBank format data format, annotation is easy to accumulate with DNannotator. It is very simple to merge annotation from different resources or from batches of annotation of the same target sequence. Its tab-delimited feature table also makes the management of annotation data convenient for both sophisticated and unsophisticated users, and can be used for sequence data extraction, providing a basis for high-throughput experimental design.
To facilitate data exchange, DNannotator provides a function that converts a native DNannotator features table into GFF (Gene-Finding Format), which is used as one of the annotation components in DAS. Therefore, annotation products for DNannotator could be stored, viewed and distributed in DAS.
DNannotator can be accessed at http://sky.bsd.uchicago.edu/DNannotator.htm. All assemblies, input and output data of annotation on 13q can be accessed through our website at http://sky.bsd.uchicago.edu/example_data.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENT
We would like to thank Hong Xu (Center for Human Genetics, Duke University Medical Center) for valuable discussion and suggestions and Jim Kent (UCSC) for help on BLAT-related issues. This work was supported by a Young Investigator Grant to C.L. and a Distinguished Investigator Grant to E.S.G. from NARSAD (National Alliance for Research in Schizophrenia and Affective Disorders) and NIH R01 MH65560-01 and R01MH59535 to E.S.G. Support from the Geraldi Norton Memorial Corporation, the Eklund Family and Anita Kaskel Roe are each gratefully acknowledged.
REFERENCES
- 1.Ouzounis C.A. and Karp,P.D. (2002) The past, present and future of genome-wide re-annotation. Genome Biol., 3, COMMENT 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camus J.C., Pryor,M.J., Medigue,C. and Cole,S.T. (2002) Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology, 148, 2967–2973. [DOI] [PubMed] [Google Scholar]
- 3.Carter K., Oka,A., Tamiya,G. and Bellgard,M.I. (2001) Bioinformatics issues for automating the annotation of genomic sequences. Genome Inform. Ser. Workshop Genome Inform., 12, 204–211. [PubMed] [Google Scholar]
- 4.Harris N.L. (1997) Genotator: a workbench for sequence annotation. Genome Res., 7, 754–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makalowska I., Ryan,J.F. and Baxevanis,A.D. (2001) GeneMachine: gene prediction and sequence annotation. Bioinformatics, 17, 843–844. [DOI] [PubMed] [Google Scholar]
- 6.Bailey L.C. Jr, Fischer,S., Schug,J., Crabtree,J., Gibson,M. and Overton,G.C. (1998) GAIA: framework annotation of genomic sequence. Genome Res., 8, 234–250. [DOI] [PubMed] [Google Scholar]
- 7.Jareborg N. and Durbin,R. (2000) Alfresco—a workbench for comparative genomic sequence analysis. Genome Res., 10, 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glusman G. and Lancet,D. (2000) GESTALT: a workbench for automatic integration and visualization of large-scale genomic sequence analyses. Bioinformatics, 16, 482–483. [DOI] [PubMed] [Google Scholar]
- 9.Taudien S., Rump,A., Platzer,M., Drescher,B., Schattevoy,R., Gloeckner,G., Dette,M., Baumgart,C., Weber,J., Menzel,U. et al. (2000) RUMMAGE—a high-throughput sequence annotation system. Trends Genet., 16, 519–520. [DOI] [PubMed] [Google Scholar]
- 10.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 11.Florea L., Hartzell,G., Zhang,Z., Rubin,G.M. and Miller,W. (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res., 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mott R. (1997) EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput. Appl. Biosci., 13, 477–478. [DOI] [PubMed] [Google Scholar]
- 13.Wheelan S.J., Church,D.M. and Ostell,J.M. (2001) Spidey: a tool for mRNA-to-genomic alignments. Genome Res., 11, 1952–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuler G.D. (1998) Electronic PCR: bridging the gap between genome mapping and genome sequencing. Trends Biotechnol., 16, 456–459. [DOI] [PubMed] [Google Scholar]
- 15.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.A. and Barrell,B. (2000) Artemis: sequence visualization and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 16.Benson D.A., Boguski,M.S., Lipman,D.J. and Ostell,J. (1997) GenBank. Nucleic Acids Res., 25, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Detera-Wadleigh S.D., Badner,J.A., Berrettini,W.H., Yoshikawa,T., Goldin,L.R., Turner,G., Rollins,D.Y., Moses,T., Sanders,A.R., Karkera,J.D. et al. (1999) A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc. Natl Acad. Sci. USA, 96, 5604–5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C., Badner,J.A., Christian,S.L., Guroff,J.J., Detera-Wadleigh,S.D. and Gershon,E.S. (2001) Fine mapping supports previous linkage evidence for a bipolar disorder susceptibility locus on 13q32. Am. J. Med. Genet., 105, 375–380. [DOI] [PubMed] [Google Scholar]
- 21.Badner J.A. and Gershon,E.S. (2002) Regional meta-analysis of published data supports linkage of autism with markers on chromosome 7. Mol. Psychiatry, 7, 56–66. [DOI] [PubMed] [Google Scholar]
- 22.Blouin J.L., Dombroski,B.A., Nath,S.K., Lasseter,V.K., Wolyniec,P.S., Nestadt,G., Thornquist,M., Ullrich,G., McGrath,J., Kasch,L. et al. (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nature Genet., 20, 70–73. [DOI] [PubMed] [Google Scholar]
- 23.Brzustowicz L.M., Honer,W.G., Chow,E.W., Little,D., Hogan,J., Hodgkinson,K. and Bassett,A.S. (1999) Linkage of familial schizophrenia to chromosome 13q32. Am. J. Hum. Genet., 65, 1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.