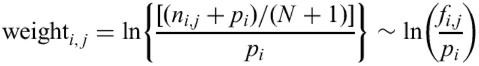Abstract
With the increasing number of eukaryotic genomes available, high-throughput automated tools for identification of regulatory DNA sequences are becoming increasingly feasible. Several computational approaches for the prediction of regulatory elements were recently developed. Here we combine the prediction of clusters of binding sites for transcription factors with context information taken from genome annotations. Target Explorer automates the entire process from the creation of a customized library of binding sites for known transcription factors through the prediction and annotation of putative target genes that are potentially regulated by these factors. It was specifically designed for the well-annotated Drosophila melanogaster genome, but most options can be used for sequences from other genomes as well. Target Explorer is available at http://trantor.bioc.columbia.edu/Target_Explorer/
INTRODUCTION
The sequencing of several eukaryotic genomes during the last decade opens new opportunities for the understanding of gene function and regulation. While significant progress has been achieved in gene prediction and functional annotation, the ability to identify regulatory elements required for the correct expression of genes is limited. The development of user-friendly computational approaches for the prediction of regulatory elements is an important goal due to the labor-intensive nature of existing wet-biology methods.
Gene regulatory elements consist of short conserved binding sites for specific transcription factors (TFs) that control the levels of gene expression in specific cell types. Differential expression of target genes that are regulated by a particular TF can be achieved by having binding sites with different but similar sequences and, therefore, different affinities for the protein. Owing to the intrinsic sequence variability of TF binding sites they must be represented by a model that summarizes information about their alignment. The simplest method for describing binding sites is the IUPAC (International Union of Pure and Applied Chemistry) consensus sequence, which indicates the predominant nucleotide or nucleotide combinations at each position in a set of training sequences (1). However, while it is easy to write a consensus sequence, it is difficult to find one that is optimal for predicting new sites. An alternative to consensus sequences is a weight matrix representation of the sites (2). Positional weight matrices store the frequency of each nucleotide at every position of the motif. The score for any particular site is calculated as the sum of matrix values for that site's sequence. Any sequence that differs from the consensus sequence will have a reduced score whose value depends on its extent of deviation from the consensus. This is a convenient way to account for the fact that some positions are more conserved than others, and presumably are more important for the activity of the site. Collections of binding site matrices are compiled in the TRANSFAC database (3). There are a number of methods for predicting new binding sites based on these libraries, for example, MatInspector (4) and MATRIX SEARCH (5).
The short length and degenerate nature of TF binding sites lead to a large number of false-positive and biologically non-functional predictions for single TFs. However, another hallmark of eukaryotic regulatory elements is that binding sites are often organized into functional groups called modules (6) where TFs bind to promoter regions and regulate transcription as a synergistic (cooperative) or antagonistic complex. Having information about combinations of TFs and their preferred positioning relative to each other can lead to a more accurate prediction of novel regulatory regions. The FastM approach together with ModelInspector (7) allows the generation of models with two TF binding sites by simply selecting them from the TRANSFAC library and using predefined models to scan sequences. Cister is another program that detects regulatory regions by searching for clusters of binding sites based on a hidden Markov model (8). Further development of tools to identify target genes have included the use of context information taken from gene sequence annotation together with TF binding site prediction. Cis-analist (9) and FlyEnhancer (10) were developed during the last year for this purpose for the Drosophila melanogaster genome. Although they are very useful tools, there are some disadvantages and restrictions in both methods (for example, users cannot create custom libraries of binding sites, FlyEnhancer uses IUPAC consensus sequences to represent binding sites and the number of identified clusters is restricted in cis-analist).
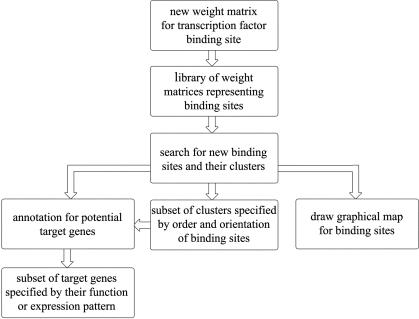
Here we present a new tool called Target Explorer, which has a user-friendly self-explanatory web interface that allows the user to: (a) create customized libraries of TF binding site matrices based on user-defined sets of training sequences; (b) search for clusters of binding sites for specified sets of TFs; and (c) extract annotation for potential target genes regulated by a specified set of TFs (Fig. 1). Target Explorer was specifically designed for the well-annotated D.melanogaster genome, and therefore accommodates searches of the entire or user-defined subsets of the genome. However, most options can also be used for sequences from other genomes.
Figure 1.
Flowchart from the Target Explorer home page depicting its functions. In order to begin a particular type of search or analysis one must click on the corresponding button. The user can start by making new matrices representing TF binding sites or choose existing matrices from the library and start a search for new binding sites or their clusters. The resulting list of putative binding sites can be translated into a graphical map, a subset of binding site clusters can be specified based on order and orientation of binding sites and flanking sequences for each cluster can be retrieved to facilitate the cloning of a cluster. Target Explorer can identify genes located near each binding site cluster and choose a subset of these genes specified by their function or expression pattern.
SOFTWARE DESCRIPTION
Generation of weight matrices
Target Explorer allows users to create customized libraries of weight matrices representing binding sites for transcription factors. Therefore, the ability to predict new binding sites does not depend on any predefined library. One can use experimental data about binding specificity for the transcription factor of interest (for example, DNaseI footprinting data or EMSA) to generate a new weight matrix and search for potential binding sites. Sets of DNA sequences of various lengths believed to contain binding sites are used as an input. They are first aligned so as to distinguish conserved binding sites using a ‘consensus’ program for local multiple sequence alignment (11). Candidate alignments are sorted by their information content (11), and the user can specify the number of alignments to observe and to choose from. The selected alignment is translated into a weight matrix using the expression:
where N is the total number of sequences in the alignment, ni, j is the number of times nucleotide i is observed in position j of the alignment, fi, j=ni, j/N is the frequency of letter i at position j, pi is the a priori probability of letter i (for example, overall frequency of letter i in the D.melanogaster genome). A positive weighti, j implies that the frequency of letter i at position j of the alignment is higher than the a priori probability of this letter. Target Explorer design also allows the editing of weights based on available mutation data. Thus, sequences with substitutions that significantly reduce TF binding efficiency can be excluded from the list of candidate binding sites by setting negative weights for ‘forbidden’ nucleotides. New matrices can be saved in the public library or in the user's private domain.
Search for candidate target genes for transcription factors and groups of factors
To start a new search for target genes for an individual transcription factor or a group of factors one must choose those matrices that represent the corresponding binding sites from the library and define the cut-off score for each matrix. The recommended cut-off score for an individual matrix is equivalent to the lowest score observed in the corresponding training set. In order to define a cluster of TF binding sites, the user must specify the minimal required number of sites per cluster for each transcription factor and the maximal length of a DNA fragment that contains all these sites. The score for an entire cluster is calculated as a sum of individual scores for the minimal required number of sites for each TF. The cut-off score for an entire cluster is taken as the sum of cut-off scores for individual sites. Because each site score is proportional to its length, scores for individual sites are normalized according to the maximal possible score for their matrices. Therefore, the maximal score for a cluster is equal to the minimal required number of sites in the cluster. The program Patser (11) was implemented in order to score individual potential binding sites against the matrix.
Cluster searches can be carried out for specified D.melanogaster sequences such as gene(s), single chromosome arms, specified cytological regions or the whole genome (Release 3.1). In addition, any sequence in fasta format can also be analysed. Search results are represented as a list of clusters that can be transformed into a graphical map where each site is depicted along the sequence line according to its position, orientation and score (see Fig. 2 for example). Subsets of clusters with specified orders and orientations of individual sites can be further selected. Flanking sequences for each cluster can be retrieved to facilitate their cloning for experimental analysis.
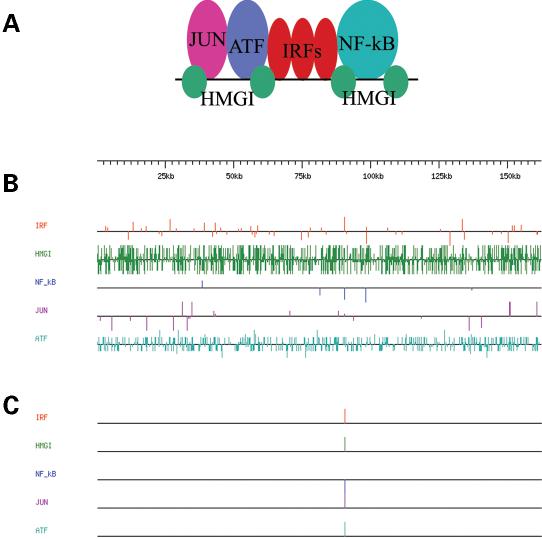
Figure 2.
Search for regulatory elements for human interferon-beta gene. (A). Diagram of the interferon-beta enhancer with transcription factors c-Jun, ATF-2, IRF, NF-kappaB and HMG I (14). (B). Example of Target Explorer graphical output for search of individual binding sites in human DNA sequence containing the interferon-beta gene (AL390882). Cut-off scores for individual matrices representing binding sites are 6.73 for c-Jun, 4.37 for ATF-2, 7.50 for IRF, 9.02 for NF-kappaB, 4.36 for HMG I. Score matrices can be found in the public domain of the Target Explorer library. Each horizontal line represents sequence and colored vertical lines represent single binding sites. The length of vertical lines is proportional to the score of the predicted binding site and the position of a line above or below the sequence line shows its orientation. (C). Target Explorer output for a search using the same parameters as in (B) but instead of searching for individual sites we searched for clusters 50 bp long that contain at least one site for each transcription factor. Target Explorer revealed only one such a cluster, which corresponds to the experimentally identified promoter for the human interferon-beta enhancer.
As a next step toward the prediction of candidate target genes Target Explorer identifies genes located near each binding site cluster. Annotation of the whole D.melanogaster genome sequence and detailed annotations for each gene are retrieved from the Fly Base, a database of the Drosophila genome (12). Based on this annotation a subset of genes can be selected that perform specific molecular functions, participate in a certain biological processes or demonstrate specific patterns of expression. A vocabulary of molecular functions, biological processes and expression patterns was obtained from the Gene Ontology Consortium (13).
FUTURE DEVELOPMENT
As the whole genome sequence of a second Drosophila species Drosophila pseodoobscura will soon become available we have begun to implement this information into Target Explorer searches for clusters of regulatory elements. Assuming that biologically significant sequences are likely to be conserved between these two Drosophila species, this comparison can further reduce the number of false-positive clusters by requiring that they exist in both species.
Although Target Explorer allows searches for clusters of binding sites in any sequence of interest, the user cannot access annotations for these clusters even if an annotation for the organism of interest already exists. We are planning to incorporate genome sequences and genome annotations for other organisms as they become available in unified formats.
AVAILABILITY
Target Explorer is accessible via the WWW interface at http://trantor.bioc.columbia.edu/Target_Explorer. The detailed manual can be found at http://trantor.bioc.columbia.edu/Target_Explorer/manual.html. For reporting problems and for asking questions emails should be sent to as1689@columbia.edu. We kindly ask that this paper be cited when results are published based on Target Explorer searches. Target Explorer has been available for public use since March 2002 and has 95 registered users as of March 2003.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jill Wildonger and Barbara Noro for helpful discussions and extensive tests during Target Explorer development. We also thank Alan Wong for his help in initiation of this project. This work was supported by NSF grant #DBI-9904841 to B.H. and by an NIH grant to R.S.M.
REFERENCES
- 1.Day W.H. and McMorris,F.R. (1992) Critical comparison of consensus methods for molecular sequences. Nucleic Acids Res., 20, 1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stormo G.D. (2000) DNA binding sites: representation and discovery. Bioinformatics, 16, 16–23. [DOI] [PubMed] [Google Scholar]
- 3.Heinemeyer T., Wingender,E., Reuter,I., Hermjakob,H., Kel,A.E., Kel,O.V., Ignatieva,E.V., Ananko,E.A., Podkolodnaya,O.A., Kolpakov,F.A. et al. (1998) Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res., 26, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q.K., Hertz,G.Z. and Stormo,G.D. (1995) MATRIX SEARCH 1.0: a computer program that scans DNA sequences for transcriptional elements using a database of weight matrices. Comput. Appl. Biosci., 11, 563–566. [DOI] [PubMed] [Google Scholar]
- 6.Werner T. (1999) Models for prediction and recognition of eukaryotic promoters. Mammal. Genome, 10, 168–175. [DOI] [PubMed] [Google Scholar]
- 7.Frech K. (1997) A novel method to develop highly specific models for regulatory units detects a new LTR in GenBank which contains a functional promoter. J. Mol. Biol., 270, 674–687. [DOI] [PubMed] [Google Scholar]
- 8.Frith M.C., Hansen,U. and Weng,Z. (2001) Detection of cis-element clusters in higher eukaryotic DNA. Bioinformatics, 17, 878–889. [DOI] [PubMed] [Google Scholar]
- 9.Berman B.P., Nibu,Y., Pfeiffer,B.D., Tomancak,P., Celniker,S.E., Levine,M., Rubin,G.M. and Eisen,M.B. (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA, 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markstein M., Markstein,P., Markstein,V. and Levine,M.S. (2002) Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 99, 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hertz G.Z. and Stormo,G.D. (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics, 15, 563–577. [DOI] [PubMed] [Google Scholar]
- 12.Ashburner M. and Drysdale,R. (1994) FlyBase—the Drosophila genetic database. Develop. Suppl., 120, 2077–2079. [DOI] [PubMed] [Google Scholar]
- 13.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yie J., Merika,M., Munshi,N., Chen,G. and Thanos,D. (1999) The role of HMG I(Y) in the assembly and function of the IFN-beta enhanceosome. EMBO J., 18, 3074–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]