Abstract
Functional annotation is routinely performed for large-scale genomics projects and databases. Researchers working on more specific problems, for instance on an individual pathway or complex, also need to be able to quickly, completely and accurately annotate sequences. The Bioverse sequence annotation server (http://bioverse.compbio.washington.edu) provides a web-based interface to allow users to submit protein sequences to the Bioverse framework. Sequences are functionally and structurally annotated and potential contextual annotations are provided. Researchers can also submit candidate genomes for annotation of all proteins encoded by the genome (proteome).
FEATURES AND IMPLEMENTATION
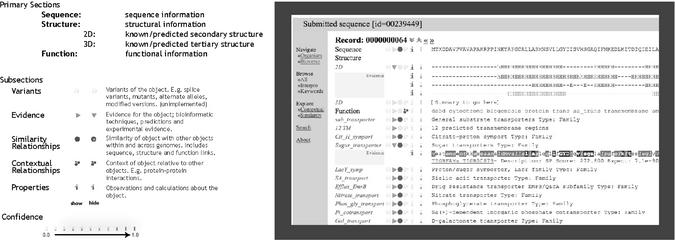
Proteomes submitted to the Bioverse annotation server are annotated using the Bioverse Action pipeline and returned as a set of Bioverse records corresponding to each protein in the proteome. Individual protein sequences submitted are compared against all sequences in Bioverse and the records for the matching sequences are returned (including the Bioverse record for the sequence itself, if the organism's proteome has been processed by Bioverse). The format of an example matching record that is returned is shown in Figure 1, with sections pertaining to each type of annotation performed outlined. The record is hierarchically organized and each section is expandable into subsections by clicking on the appropriate icon next to the section name. Some features of the annotation are detailed below.
Figure 1.
Screen shot of the Bioverse record interface showing the main sections with some expanded subsections. Sequence-based data is displayed horizontally starting with the first residue in the style of a sequence alignment. Confidence values are assigned to objects based on the evidence available for that object.
The sequence section lists similar sequences identified by searches of both the NCBI non-redundant sequence database and the Bioverse database [performed using a variety of methods, including PSI-BLAST (1)]. Matching sequences are displayed aligned with the submitted sequence, along with confidence scores and links to the original data sources.
The structure section is composed of two subsections: secondary and tertiary structure information. The secondary structure subsection shows an overall prediction combined from several sources (2–4). Expanding this subsection's ‘Evidence’ link will display the information used in making the overall prediction. Secondary structure evidence used currently includes alignments to proteins of known structure, i.e. the Protein Data Bank [PDB (2)], neural-network based secondary-structure prediction methods (4) and transmembrane region prediction (3). These data sources are combined using an artificial neural network (ANN), resulting in an overall prediction as well as a confidence measure that is derived from its output. After training with a known data set, the method outperformed each of the individual prediction methods and can be easily adapted to serve as a model for other kinds of data integration.
The tertiary structure evidence section currently includes matching sequences with known structures. The function section combines a number of different methods and databases to match sequences to patterns [PROSITE (5), BLOCKS (6), PRINTs (7)] or domains and families [Pfam (8), ProDom (9), SMART (10), TIGRFAMs (11)]. These sources are then combined to provide Interpro (12) and GO (13) categories which provide the primary annotations.
Contextual information is provided by comparing submitted sequences to a database of experimentally-derived protein–protein interactions compiled from several sources [including the Database of Interacting Proteins (DIP) (14) and the PDB]. Predicted interaction partners are listed under the Function-Context section. When applied to complete proteomes, networks of interacting proteins can be extracted and matched to metabolic and regulatory pathways [such as KEGG (15)]. These networks can be interactively explored on the Bioverse website.
FUTURE WORK
The tertiary structure evidence section will be expanded to include three-dimensional structural prediction using comparative de novo modeling techniques developed by our group (16). Besides protein–protein interactions, the contextual information section will incorporate protein and gene expression data to provide a more comprehensive picture of the proteome.
CONCLUSIONS
Bioverse is a valuable tool for annotation of proteins and proteomes. Sequence, structural, functional and contextual annotation is performed and results in each section are integrated. Bioverse allows researchers to submit sequences for complete annotation, explore completed proteomes, interactively browse contextual networks of proteins and perform queries on these proteomes. Applications of Bioverse include whole genome annotation, protein complex characterization, study of host–pathogen interactions and hypothesis generation for proteins of unknown function. The Bioverse database and annotation tool is available at (http://bioverse.compbio.washington.edu).
CALCULATION TIMES AND CURRENT USAGE
A proteome consisting of up to 15 000 proteins takes <1 day to be processed by us. Individual searches are returned within seconds or minutes. The web server currently receives ∼4000 unique visitors each month, resulting in >12 000 ‘hits’ and >1500 queries/searches.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by a Searle Scholar Award and NSF Grant DBI-0217241 to R.S. and the University of Washington's Advanced Technology Initiative in Infectious Diseases.
REFERENCES
- 1.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 2.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeda M., Arai,M., Lao,D.M. and Shimizu,T. (2002) Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol., 2, 19–33. [PubMed] [Google Scholar]
- 4.Jones D.T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol., 292, 195–202. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attwood T.K., Flower,D.R., Lewis,A.P., Mabey,J.E., Morgan,S.R., Scordis,P., Selley,J.N. and Wright,W. (1999) PRINTS prepares for the new millennium. Nucleic Acids Res., 27, 220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corpet F., Gouzy,J. and Kahn,D. (1998) The ProDom database of protein domain families. Nucleic Acids Res., 26, 323–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letunic I., Goodstadt,L., Dickens,N.J., Doerks,T., Schultz,J., Mott,R., Ciccarelli,F., Copley,R.R., Ponting,C.P. and Bork,P. (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res., 30, 242–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haft D.H., Loftus,B.J., Richardson,D.L., Yang,F., Eisen,J.A., Paulsen,I.T. and White,O. (2001) TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res., 29, 41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2000) InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics, 16, 1145–1150. [DOI] [PubMed] [Google Scholar]
- 13.The Gene Ontology Consortium (2001) Creating the gene ontology resource: design and implementation. Genome Res., 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xenarios I., Salwinski,L., Duan,X.J., Higney,P., Kim,S.M. and Eisenberg,D. (2002) DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res., 30, 303–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M., Goto,S., Kawashima,S. and Nakaya,A. (2002) The KEGG databases at GenomeNet. Nucleic Acids Res., 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samudrala R. and Levitt,M. (2002) A comprehensive analysis of 40 blind protein structure predictions. BMC Struct Biol., 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]