Abstract
The residue-based diagram editor (RbDe) is web-based software that greatly simplifies the construction of schematic diagrams of proteins. Residue-based diagrams display the sequence of a given protein in the context of its secondary and tertiary structure. Such diagrams are frequently used to summarize mutations or sequence features, in the context of the overall topology of a protein. The initial version of RbDe was designed for transmembrane proteins and has enabled many users to create diagrams of large systems such as G protein-coupled receptors or transporters. We present an extended diagram editor that supports other families of proteins. Users can now import custom-diagram layouts, use them to render members of any protein family and generate high-quality output for publication purposes. RbDe is available free over the web, at http://icb.mssm.edu/crt/RbDe
INTRODUCTION
Residue-based diagrams of proteins, also called snake-like diagrams or protein plots, are 2D representations of a protein sequence that contains information about properties such as secondary structure. Figure 1 shows an example of a diagram drawn with the first release of RbDe. Since 1999, we have been developing and maintaining the residue-based diagram editor (RbDe) to help users create residue-based diagrams of transmembrane proteins interactively. RbDe was designed from the ground up as a web application and has been available freely over the web since its initial release (1,2).
Figure 1.
Diagram of the human dopamine transporter. The diagram was produced through the web with the first release of RbDe. Residues are colored according to residue types (the color scheme colors residues of similar physicochemical properties in a similar way) and the diagram indicates the element of secondary structure (transmembrane helices) and the connecting loops. The white circles represent contiguous stretches of residues which are omitted from this diagram.
Here we present a new release of RbDe that supports the interactive creation of diagrams with custom layout.
RELATED WORK
Several programs have been developed to automate the rendering of diagrams for membrane proteins or assist in their drawing. Among these, Viseur (http://transport.physbio.mssm.edu/viseur) (3,4), VHMPT (5) and RbDe, have focused on allowing users to construct diagrams interactively and to adjust the parameters that control the appearance of a diagram through a graphical user interface. TEXtopo (6) focused on creating highly customizable diagrams that can produce high quality output for publication. High quality output could also be obtained with the Viseur program when using the encapsulated postscript output, but customization options were very limited in this program (Viseur was designed mostly for proteins in the G protein-coupled receptor family). TEXtopo allows users to place labels on the diagram, add legend and graphical annotations to certain residues and control to some extent the appearance of loops. In order to use TEXtopo, users must be familiar with the LaTeX typesetting system (7). This is a factor that probably limits the applicability of this tool.
RbDe
In contrast, RbDe was designed and implemented as a web application and can be accessed from most web browsers. Figure 2 illustrates how a user progresses through the software to create a diagram. Since it was originally published in 2000 (2), RbDe has been used to create an average of five diagrams a day by registered users from over 40 countries.
Figure 2.
Progression of a typical user through the web application for a typical use of RbDe. Information is collected as the user progresses through the sections of the web software (seq: sequence; ss: secondary structure; layout: layout of the diagram on the page, see text for details; colors: colors of specific residues or color scheme to color the entire diagram by residue type; URLs: hyperlinks from residues to documents on the web). The diagram is built and refined iteratively as the user proceeds through the application.
CUSTOM LAYOUTS
In the new extended version of RbDe, users can import a custom diagram layout. The layout determines how sequences imported in RbDe will be laid out in the final picture. In contrast to the first version of RbDe, the extended version presented here makes it possible to customize the overall placement of the secondary structure elements in the diagram. This feature allows users to customize layouts for a specific purpose, such as the rendering of diagrams for a specific family of proteins, or to refine the presentation of existing diagrams of transmembrane proteins.
RbDe accepts layouts encoded as described for the residue-based diagram generator (RbDg) (8). The creation of a layout input file for RbDe requires knowledge of XML and a few conventions described in the RbDg XML schema documentation (see http://icb.mssm.edu/crt/RbDg). The development of a new diagram layout can be performed in an iterative manner, usually by trial and error, after an outline of the final layout has been created on paper. Figure 3 shows a fragment of an input file, and the corresponding fragment in the diagram produced by RbDe, as the layout directives are interpreted to render a sequence.
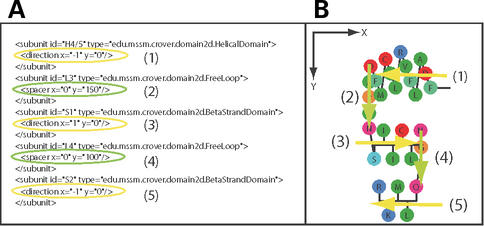
Figure 3.
(A) Fragment of a layout input file. A layout is defined by a list of subunits. Each subunit has a type, which determines how the residues in the subunit will be rendered. Some subunit types (e.g. helices and strands) are given a direction. The direction is a vector that controls the global direction along which residues of the subunit will be rendered. Other subunits, such as subunits to represent loops that connect the first type of subunits, are given a spacer. The spacer is a vector that defines how much the first residue of the subunit is separated from the last. Units of direction and spacer are pixels in the coordinate system shown on the right. (B) Fragment of a diagram built with the layout parameters shown on the left. Yellow arrows represent the direction of helices and strands. Green arrows represent the spacers of the loops.
SCALABLE VECTOR GRAPHICS OUTPUT
In addition to image outputs, RbDe can generate diagrams in the scalable vector graphics (SVG) format (http://www.w3.org/TR/SVG/). SVG is a vectorial format that is gaining growing acceptance as a standard for exchange of high-quality images, schemas and diagrams on the web. Diagrams produced with RbDe can be exported to SVG by clicking on a link in the diagram library (see menu on the top of the application screens). The resulting SVG document is visualized with an appropriate plug-in, and/or can be saved to disk. The SVG document can be read in Adobe Illustrator (version 10+) or similar tools with SVG support. Since SVG is a vectorial format, parts of the figure can be scaled, removed, re-colored or altered in a variety of ways, to further customize the diagram, for instance for publication purposes. (Diagrams shown in Figs 1, 3 and 4 were generated with RbDe and edited with Adobe Illustrator 10.)
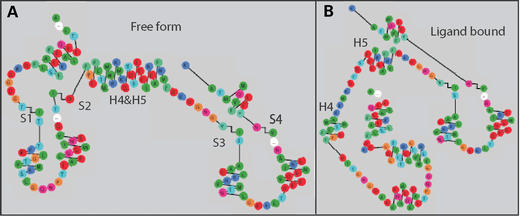
Figure 4.
The calmodulin protein sequence is rendered and shown with two diagram layouts based on the crystal structure of the molecule (A) and the calmodulin-trifluoperazine complex (B). The diagrams reveal the structural rearrangement upon ligand binding.
APPLICATION
As an example of custom layout, we have built a diagram of the rat calmodulin protein and its complexed form with trifluoperazine. The diagrams were constructed in the following manner: the secondary structure elements were determined with the Kabsch and Sander algorithm (9) from crystal structure in the Protein Data Bank (PDB ID: 3CLN and 1A29). Visual inspection of the 3D structures suggested an arrangement for the secondary structure elements in the 2D diagram. The layouts were further adjusted to eliminate superposition of residues. An RbDg input file was then built iteratively for each conformation of the protein, using as a guide the initial sketch, and visualizing the diagram after adding each helix or strand. Creating a diagram layout is possible over the web, but for faster iterations we recommend using a local installation of the diagram generator. The final layout portion of the RbDg input file can then be uploaded to RbDe and used to plot proteins that share the same secondary structure.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants P01 DA124080, P20 HL69733 and the Institute for Computational Biomedicine at MSSM.
REFERENCES
- 1.Campagne F., Jestin,R., Reversat,J.L., Bernassau,J.M. and Maigret,B. (1999) Visualisation and integration of G protein-coupled receptor related information help the modelling: description and applications of the Viseur program. J. Comput. Aided Mol. Des., 13, 625–643. [DOI] [PubMed] [Google Scholar]
- 2.Konvicka K., Campagne,F. and Weinstein,H. (2000) Interactive construction of residue-based diagrams of proteins: the RbDe web service. Protein Eng., 13, 395–396. [DOI] [PubMed] [Google Scholar]
- 3.Campagne F. (1995) The Viseur Program. http://transport.physbio.mssm.edu/viseur. Laboratoire de Chimie Theorique, Nancy, France.
- 4.Campagne F. and Weinstein,H. (1999) Schematic representation of residue-based protein context-dependent data: an application to transmembrane proteins. J. Mol. Graph. Model., 17, 207–213. [DOI] [PubMed] [Google Scholar]
- 5.Lin W.J. and Hwang,M.J. (1998) VHMPT: a graphical viewer and editor for helical membrane protein topologies. Bioinformatics, 14, 866–868. [DOI] [PubMed] [Google Scholar]
- 6.Beitz E. (2000) T(E)Xtopo: shaded membrane protein topology plots in LAT(E)X2epsilon. Bioinformatics, 16, 1050–1051. [DOI] [PubMed] [Google Scholar]
- 7.Lamport L. (1994) LaTeX: A Document Preparation System, 2nd Ed., Addison Wesley. [Google Scholar]
- 8.Campagne F., Bettler,E., Vriend,G. and Weinstein,H. (2003) Batch mode generation of residue-based diagrams of proteins. Bioinformatics, in press. [DOI] [PubMed] [Google Scholar]
- 9.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]