Abstract
Protein structure prediction is a cornerstone of bioinformatics research. Membrane proteins require their own prediction methods due to their intrinsically different composition. A variety of tools exist for topology prediction of membrane proteins, many of them available on the Internet. The server described in this paper, BPROMPT (Bayesian PRediction Of Membrane Protein Topology), uses a Bayesian Belief Network to combine the results of other prediction methods, providing a more accurate consensus prediction. Topology predictions with accuracies of 70% for prokaryotes and 53% for eukaryotes were achieved. BPROMPT can be accessed at http://www.jenner.ac.uk/BPROMPT.
INTRODUCTION
Membrane proteins are vital cellular components (1) and their prediction is a cornerstone of bioinformatics research. Alpha-helical membrane proteins are responsible for the majority of interactions between a cell and its environment (2). The transmembrane (TM) helices are characterised by long stretches of predominantly hydrophobic residues (typically 17–25) (3) which is sufficient to cross the hydrophobic region of the lipid bilayer (2.5 nm) (4). The compositional bias for hydrophobicity arises because these residues are required to interact with the hydrophobic lipid environment of the membrane.
A number of methods exist to predict TM alpha helices employing a wide range of techniques. Methods have been devised that use the amino acid preference for membrane and non-membrane segments of proteins (5), e.g. TMpred, which uses statistical preferences to predict TM-helices taken from an expert-compiled data set of membrane proteins (6). TopPred II applies the ‘positive inside rule’ to evaluate the validity of topology models derived from hydropathy analysis (7). This method uses several different preference matrices to increase accuracy and was developed further by the SOSUI predictor (8). DAS is based on low-stringency dot-plots of the query sequence against a collection of non-homologous membrane proteins using a previously derived, special scoring matrix (9). As well as using location preference and hydropathy scales, other physicochemical parameters, such as protein length and charge, were used to better characterise TM domains. Currently, the best performing alpha-helical predictors, TMHMM2 (10) and HMMTOP2 (11), are based on hidden Markov models (HMM) that model a variety of constraints on membrane protein structure caused by the lipid bilayer.
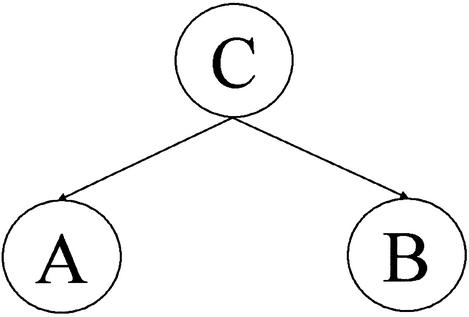
A Bayesian Belief Network (BBN) is a probabilistic model consisting of a directed graph, together with an associated set of probability tables (12,13). The graph consists of nodes and arcs as shown in Figure 1. The nodes represent variables which can be discrete or continuous. The arcs represent causal/influential relationships between variables. Variable A is conditionally independent from B given C if P(A, B|C)=P(A|C)P(B|C) or equivalently, P(A|B, C)=P(A|C) where the notation P(Y|X) denotes the probability of Y given X. Using these conditional dependencies, the joint probabilities of all the variables in the model can be factored into a product of conditional probabilities. For example P(A, B, C)=P(C)P(A|C)P(B|C).
Figure 1.
A schematic example of a simple Bayesian Network showing three nodes: one parent and two daughters.
Bayesian network probabilistic models provide a flexible and powerful framework for statistical inference and learn model parameters from data (14). The goal of inference is to find the distribution of a random variable in the network conditioned on values of other variables in the network. BBNs can be used to efficiently estimate optimal values of model parameters from data. Another major advantage of BBNs is the ability to combine machine learning with expert opinions. Weights and/or causal relationships can be specified before network training occurs. This allows relationships to be represented that are known to be true or to be forbidden if they can never occur.
MATERIALS AND METHODS
This paper presents an internet server that implements a consensus method for predicting alpha-helical membrane protein topology. Predictions are obtained from a range of web-based predictors and are combined using a BBN.
Test and training data
In order to properly benchmark this method against the individual methods from which it is built, the test set assembled by Ikeda et al. (15) was used. This paper describes an independent test of prediction accuracy for all the individual servers used and therefore, using the same test set, provides a way of rating the true accuracy of BPROMPT. The test set used contains 52 eukaryotic and 70 prokaryotic proteins with experimentally derived topologies. This is a non-redundant dataset where similarity between sequences is <30% in all cases.
In order to evaluate the ability of BPROMPT to discriminate TM and non-TM proteins, a second test set was compiled. This was a set of 591 known cytoplasmic or periplasmic soluble proteins obtained from SWISS-PROT release 41.0 (18).
The training set was compiled from two sources: the database compiled by Möller et al. (16) and the MPtopo database (17). Topologies obtained from the Möller database corresponded to proteins for which reliable experimental topology information is available. For the MPtopo database, only proteins where either the three-dimensional structure has been determined or where the approximate position of TM helices has been determined experimentally using gene fusion, proteolytic fusions or some other biochemical characterisation. If a protein was present in both databases, the Möller database entry was used. This gave a training set of 124 proteins. Any sequences from the test set that were present in the training set were removed from the training set.
Consensus transmembrane topology prediction
The predictors used are HMMTOP2, DAS, SOSUI, TMpred and TopPred II (for web site addresses see Table 1). Predictions for the training set were obtained from each predictor and the results saved. A BBN was constructed consisting of six nodes, five evidence nodes (one for each of the predictors) and a decision node. There is a direct causal relationship from every evidence node to the final decision node. The network was then trained by comparing each prediction with the known structure, allowing the BBN to learn how to identify the strengths and weaknesses of each method.
Table 1. Web site addresses of individual methods used in BPROMPT.
| Predictor | Web site address |
|---|---|
| HMMTOP2 | http://www.enzim.hu/hmmtop/ |
| DAS | http://www.sbc.su.se/~miklos/DAS/ |
| SOSUI | http://sosui.proteome.bio.tuat.ac.jp/sosuiframe0.html |
| TMpred | http://www.ch.embnet.org/software/TMPRED_form.html |
| TopPred II | http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html |
A web page was constructed to act as an interface to the Perl CGI server. Once a sequence has been entered, it is sent to each web server, where its structure is predicted by each method. The results are returned to the interface where the predictions are parsed and passed to the BBN. The BBN decides which of the predicted TM segments are most likely to be true and then returns this to the interface where the results are displayed.
The final stage of the prediction process is post-network processing. The aim here is to alter the prediction to conform to known structural tendencies of alpha helices. To this end, any prediction shorter than 10 residues is discarded, as this is shorter than the minimum allowed length of alpha helices.
RESULTS
The accuracy of prediction was measured in two ways: the number of TM segments predicted compared to the actual number in the protein and the topology of the protein. Topology prediction is defined, in the context of this paper, as prediction of the number and location of TM regions combined with prediction of N-terminal location. The accuracies of the consensus method are summarised in Table 2. Accurate identification of a TM segment is assumed if the central residue of the predicted TM helix is within 11 residues of the position of the actual central residue of the helix, which is the accuracy measure used by Ikeda et al. (15). In order to effect an unbiased comparison of BPROMPT with the methods examined by Ikeda et al., we also use their criteria for accuracy.
Table 2. Topology prediction accuracies for the BPROMPT server, reported separately for eukaryotes and prokaryotes.
| Eukaryotic accuracies (%) | Prokaryotic accuracies (%) | ||
|---|---|---|---|
| Number of helices | Topology | Number of helices | Topology |
| 60.67 | 53.41 | 78.9 | 70.12 |
The accuracy of discrimination between TM and soluble proteins was expressed as the percentage of soluble proteins with an incorrect TM region prediction of the total 591 soluble proteins tested. Only 4.06% (24/591) of soluble proteins tested had false TM segment predictions. Only one of the 24 false positives had more than one TM region predicted. This result compares favourably with other methods. Testing of a set of soluble proteins undertaken previously (6) showed that false positive rates were typically ≥7%. The best of the methods used as part of BPROMPT was SOSUI which had an accuracy of 2.99%. However, this method also underpredicts the number of membrane proteins. It must be stressed that while the two test sets used were different, they are of comparable size and the results can be roughly equated.
DISCUSSION
The aim of this work was to provide a publicly available method with improved alpha helical transmembrane protein prediction. Our server utilises a range of web-based predictors and then combines them into a consensus prediction using a BBN. An improved accuracy in topology prediction was achieved when compared to currently available methods (Table 3). Increased accuracies of 13% for eukaryotes and 6% for prokaryotes were obtained compared to the best performing of the individual predictors (HMMTOP 2.0). Nevertheless, improvements could be made to the method to increase its sensitivity. Short predictions of the core of a helix were rejected by the post-network processing, suggesting the provision of an option to allow the overall architecture of the protein to be better visualised by also reporting such reliable, but too short, TM region predictions. However, including these short segments may be difficult to accomplish without including many more false positive predictions.
Table 3. Topology prediction accuracies of individual methods used in consensus prediction (15), reported for eukaryotes and prokaryotes separately.
| Methods | Topology prediction accuracies (%) | |
|---|---|---|
| Eukaryotic | Prokaryotic | |
| TMpred | 32.7 | 35.7 |
| TopPred II | 21.2 | 55.7 |
| DAS | 28.8* | 34.3* |
| SOSUI | 53.8* | 51.4* |
| HMMTOP 2.0 | 40.4 | 64.3 |
The method implemented in our server improves TM prediction accuracy beyond that of the individual predictors, as has been shown with other published, but not publicly available, consensus methods. Although the increase in accuracy achieved is more modest than might have been hoped, this tool nonetheless represents an important advance. With the large number of genomes either published or being sequenced, there is an increasing need to develop accurate annotation methods. 20–30% of most genomes are membrane proteins (19,20) and thus any increase in accuracy will be of great benefit in annotation efforts. Membrane proteins also often provide very fruitful therapeutic targets. The most obvious examples are the G protein-coupled receptors, which are the target of ∼50% of all marketed drugs (21). Increasing the accuracy of membrane protein topology prediction will probably facilitate the pace of drug design.
Acknowledgments
ACKNOWLEDGEMENTS
Many thanks go to Helen Kirkbride for comments and help. P.D.T. is grateful to the Medical Research Council for a priority area studentship in Bioinformatics.
REFERENCES
- 1.Efremov G., Nolde,E., Vergoten,G. and Arseniev,A. (1999) A solvent model for simulations of peptides in bilayers. Biophys. J., 76, 2248–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frishman D. and Mewes,H.W. (1997) Protein structural classes in five complete genomes. Nature Struct. Biol., 4, 626–628. [DOI] [PubMed] [Google Scholar]
- 3.von Heijne G. (1994) Membrane Protein Assembly: rules of the game. BioEssays, 17, 25–30. [DOI] [PubMed] [Google Scholar]
- 4.Deisenhofer J., Remington,S.J. and Steigemann,W. (1985) Experience with various techniques for the refinement of protein structures. Methods Enzymol., 115, 303–323. [DOI] [PubMed] [Google Scholar]
- 5.Juretic D., Lee,B., Trinajstic,N. and Williams,R.W. (1993) Conformational preference functions for predicting helices in membrane proteins. Biopolymers, 33, 255–273. [DOI] [PubMed] [Google Scholar]
- 6.Moller S., Croning,M.D.R. and Apweiler,R. (2001) Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics, 17, 646–653. [DOI] [PubMed] [Google Scholar]
- 7.Claros M.G. and von Heijne,G. (1994) TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci., 10, 685–686. [DOI] [PubMed] [Google Scholar]
- 8.Mitaku S., Ono,M., Hirokawa,T., Boon-Chieng,S. and Sonoyama,M. (1999) Proportion of membrane proteins in proteomes of 15 single-cell organisms analyzed by the SOSUI prediction system. Biophys. Chem., 82, 165–171. [DOI] [PubMed] [Google Scholar]
- 9.Cserzo M., Wallin,E., Simon,I., von Heijne,G. and Elofsson,A. (1997) Prediction of transmembrane alpha-helices in procariotic membrane proteins: the Dense Alignment Surface method. Protein Eng., 10, 673–676. [DOI] [PubMed] [Google Scholar]
- 10.Sonnhammer E.L., von Heijne,G. and Krogh,A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol., 6, 175–182. [PubMed] [Google Scholar]
- 11.Tusnady G.E. and Simon,I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics, 17, 849–850. [DOI] [PubMed] [Google Scholar]
- 12.Pearl J. (1988) Probablistic Reasoning in Intellegent Systems: Networks of Plausible Inference. Morgan Kaufman, San Mateo, California. [Google Scholar]
- 13.Cowell R.G., Dawid,A.P., Lauritzen,S.L. and Speigelhalter,D.J. (1999) Probablistic Networks and Expert Systems. Springer, New York. [Google Scholar]
- 14.Jensen F.V. (1996) Introduction to Bayesian Networks. Springer, New York. [Google Scholar]
- 15.Ikeda M., Arai,M., Lao,D.M. and Shimizu,T. (2002) Transmembrane topology prediction methods: a re-assessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol., 2, 19–33. [PubMed] [Google Scholar]
- 16.Moller S., Kriventseva,E.V. and Apweiler,R. (2000) A collection of well characterised integral membrane proteins. Bioinformatics, 16, 1159–1160. [DOI] [PubMed] [Google Scholar]
- 17.Jayasinghe S., Hristova,K. and White,S.H. (2001) MPtopo: a database of membrane protein topology. Protein Sci., 10, 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boeckmann B., Bairoch,A., Apweiler,R., Blatter M.-C., Estreicher,A., Gasteiger,E., Martin,M.J., Michoud,K., O'Donovan,C., Phan,I. et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res., 31, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallin E. and von Heijne,G. (1998) Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci., 7, 1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones D.T. (1998) Do transmembrane protein superfolds exist? FEBS Lett., 423, 281–285. [DOI] [PubMed] [Google Scholar]
- 21.Flower D.R. (1999) Modelling G-protein-coupled receptors for drug design. Biochim. Biophys. Acta, 1422, 207–234. [DOI] [PubMed] [Google Scholar]