Abstract
The fortran program ESPript was created in 1993, to display on a PostScript figure multiple sequence alignments adorned with secondary structure elements. A web server was made available in 1999 and ESPript has been linked to three major web tools: ProDom which identifies protein domains, PredictProtein which predicts secondary structure elements and NPS@ which runs sequence alignment programs. A web server named ENDscript was created in 2002 to facilitate the generation of ESPript figures containing a large amount of information. ENDscript uses programs such as BLAST, Clustal and PHYLODENDRON to work on protein sequences and such as DSSP, CNS and MOLSCRIPT to work on protein coordinates. It enables the creation, from a single Protein Data Bank identifier, of a multiple sequence alignment figure adorned with secondary structure elements of each sequence of known 3D structure. Similar 3D structures are superimposed in turn with the program PROFIT and a final figure is drawn with BOBSCRIPT, which shows sequence and structure conservation along the Cα trace of the query. ESPript and ENDscript are available at http://genopole.toulouse.inra.fr/ESPript.
INTRODUCTION
Proteins with sequence identity >30% normally belong to the same family and have similar conformation and function (1,2). Such clear homologues are likely to have diverged from a common ancestor and their sequences may show conserved differences between species of organisms. Convergent evolution can also occur within a family, if certain homologues have developed additional functions such as the capability to bind ligands away from the active site. The side function is also likely to be written in the sequence and better still in the 3D structure which is generally even more conserved. Thus, the simultaneous comparison of sequence and structure information is of importance to detect biological specificities in a group of proteins.
The program ESPript, Easy Sequencing in PostScript, generates figures of aligned sequences with secondary structure information (3). It can serve as a tool for structure/function analyses. ESPript reads text outputs from multiple sequence alignment programs such as Clustal (4) and MULTALIN (5), as well as from programs able to identify secondary structure elements from structure files such as DSSP (6) and STRIDE (7). Residues are boxed according to their similarity score and secondary structure elements are drawn at the top of sequences blocks. ESPript can be used for publication purposes and the user has access to numerous features to optimize its figure (selection of displayed sequences, choice of colours, symbols to highlight chosen residues). The program is written in Fortran and can be executed locally on Linux or Unix machines or on a web server via a CGI interface.
A server named ENDscript was later created to routinely produce ESPript figures with a maximum of sequence/structure information (8). The query is the code of a protein structure deposited with the Protein Data Bank (9) or a file with atomic coordinates. NMR and crystallographic structures in PDB format are supported. The sequence is extracted from the query and ENDscript performs a BLAST search (10) against a selected protein sequence database to detect clear homologues (search with E-value cutoff of 10−6 by default). The result is piped to Clustal or MULTALIN for a multiple sequence alignment. The final ESPript figure shows the aligned sequences with the secondary structure elements of each sequence of known 3D structure. Additional information is presented, such as intermolecular and protein–ligand contacts detected by CNS (11), accessibility calculated by DSSP and hydropathy. A BOBSCRIPT (12) figure is also generated, showing a ribbon representation of the query coloured according to sequence homology. The molecule can be rotated via a VRML file produced by MOLSCRIPT (13). Finally, a link to the program PHYLODENDRON (© 1997 by D.G. Gilbert) can be activated to draw a phylogenetic tree.
ESPript has been configured on our server so as to be used via ENDscript and to take into account the continuous increase of structures deposited with the PDB (>3000 per year) due to the development of structural genomics. The program can display up to 1000 sequences with accompanying secondary structure elements aligned on 2500 columns and 30 pages.
IMPROVEMENTS
To ESPript
Most options were available in 1998 when the reference article was published. The drawing code used in PostScript figures has been conserved since then. However, the Fortran program and the CGI script of the interface have been extensively modified, so as to enable links with other bioinformatics servers which address various aspects of the structure/sequence analysis. Connections to ESPript are now available from the protein domain database ProDom (14), the structure prediction server PredictProtein (15) and the server performing sequence analysis and similarities searches NPS@ (16). In each case, an ESPript figure with multiple sequence alignment and secondary structure elements can be generated with one or two clicks. Final main changes have been introduced with the conception of ENDscript.
The control panel of the interface has been redesigned this year. Buttons are now positioned at the top of the form (Fig. 1) and the resulting output is displayed in a full window, with all warnings, files and figures produced by ESPript immediately visible.
Figure 1.
Screen capture of the ESPript web interface: the control bar allows the user to submit its query when the main form is filled; its buttons allow swapping between form, result and log pages, to customize the input mode, and finally to load, save or quit a session.
To ENDscript
Databases necessary to ENDscript are at present downloaded and updated automatically on our server; that is, the structural database PDB, the sequence database SWISS-PROT (17), the sequence database PDBaa which is derived from the PDB and ∼100 sequenced genomes. In addition, a list of cross-reference links of sequence names between the PDB and the SWISS-PROT is downloaded monthly from the SWISS-PROT server.
The program PROFIT (Martin, A.C.R., http://www.bioinf.org.uk/software/profit) is now used in ENDscript, to superimpose known 3D structures of homologous sequences onto the query and to calculate rms deviation by Cα pairs over the fitted region. A new BOBSCRIPT figure is produced in turn, with the backbone of the query rendered as a tube, its radius being proportional to the calculated rms deviation; colour varies from white to red according to similarity (low to high). Such a figure can reveal conserved domains to the naked eye as shown below.
EXAMPLE
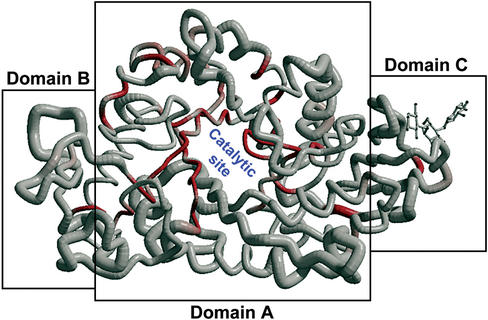
Alpha-amylases belong to the family 13 of glycoside hydrolases and hydrolyse the α-D-(1,4)-glucosidic linkages in starch-related polysaccharides (18). They generally consist of three structural domains: the conserved central domain (domain A) containing the active site, the domain B protruding from domain A and the domain C at the C-terminal region. A new binding site has been observed in the domain C of the 2 Å crystal structure of the isozyme 1 of barley α-amylase (AMY1) in complex with a thio-maltodextrin substrate analogue (19). The question arises whether the new site with no catalytic activity has biological implications. The structure was submitted to ENDscript and the search for homologues was performed against the PDBaa. One representative sequence/structure per detected organism was kept afterwards for simplification. Figure 2 was drawn with BOBSCRIPT after superimposition with PROFIT: the core appears highly conserved as shown by the red colour and the small radius of the tube; indeed it contains a beta-barrel which is a signature for hydrolases of family 13 according to the CATH classification (20). The domain C of AMY1 is short in sequence regarding the alignment produced by Clustal and rendered by ESPript (Fig. 3). The closest detected homologue is 1AMY, which is the PDB code for the other known structure of plant α-amylase. Other similar sequences are α-amylases from fungi, microbial, insects and mammalian species. Residues of AMY1 in contact with the substrate-analogue are marked by quotes on the bottom line of the figure: tyrosine 380 is conserved in the two plant α-amylases and is highly implicated in substrate binding.
Figure 2.
Cα trace of the structure of AMY1 coloured from white to red according to sequence similarities (from low to high). The radius of the Cα tube is proportional to rms deviation after superimposition of the structure of AMY1 with its homologues. The most conserved region is the beta-barrel of domain A containing the catalytic site. The thio-maltodextrine molecule bound to the domain C is shown in ball-and-sticks. Figure prepared with BOBSCRIPT (12).
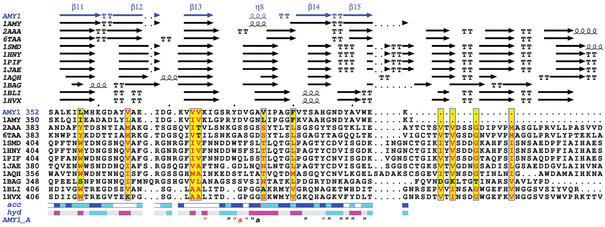
Figure 3.
ESPript output obtained from sequences homologous to AMY1. Each sequence has a known 3D structure. Secondary structure elements are presented on top: helices with squiggles, beta strands with arrows, turns with TT letters. Conserved residues are written in red in sequences block. Accessibility of AMY1 is rendered by a bar below: blue is accessible, cyan is intermediate, white is buried. Hydropathy of AMY1 is rendered by a second bar: pink is hydrophobic, cyan is hydrophilic. Contacts are shown at bottom: the letter ‘a’ points out residues making crystallographic contacts; the character “ identifies residues in contact with the thio-maltodextrin molecule; a red character at this line identifies residues having close contacts (distance<3.2 Å).
Subsequent investigations have demonstrated the role of this tyrosine in substrate recognition at this new binding site, which may enhance the efficiency of plant α-amylases over starch granules. All figures described in this section have been obtained in a few minutes with ENDscript.
FUTURE DEVELOPMENTS
Structure-based sequence alignment programs such as T-Coffee (21) will be added in the next versions of ENDscript. These programs are likely to greatly enhance the quality of the sequence alignment. For example, no insertion should be observed in strand β5 of 1AMY as displayed on the sequence alignment produced by Clustal (Fig. 3), which relies only on sequence information. However, these programs are CPU consuming and a faster ESPript/ENDscript server will be made available in autumn 2003.
AVAILABILITY
ESPript/ENDscript can be executed on a web server at http://genopole.toulouse.inra.fr/ESPript. Codes and scripts necessary to install ESPript/ENDscript are freely available for academic users and can be downloaded via ftp anonymous (ftp://ftp.toulouse.inra.fr/pub/ESPript) after completion of a licence form. A fee of 1000 Euros is required for commercial users. Licences for accompanying programs used in ENDscript (BLAST, Clustal, MULTALIN, DSSP, CNS, BOBSCRIPT, MOLSCRIPT, PHYLODENDRON and PROFIT) must be requested separately.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Richard Haser and Daniel Kahn for their support in the project. We are grateful to Robert Esnouf for modifications in BOBSCRIPT which allow the drawing of the new figure in ENDscript.
REFERENCES
- 1.Chothia C. and Lesk,A.M. (1980) How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globin. J. Mol. Biol., 136, 225–270. [DOI] [PubMed] [Google Scholar]
- 2.Chothia C. and Lesk,A.M. (1986) The relation between the divergence of sequence and structure in proteins. EMBO J., 5, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouet P., Courcelle,E., Stuart,D.I. and Metoz,F. (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- 4.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res., 16, 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 7.Frishman D. and Argos,D. (1995) Knowledge-based secondary structure assignment. Proteins, 23, 566–579. [DOI] [PubMed] [Google Scholar]
- 8.Gouet P. and Courcelle,E. (2002) ENDscript: a workflow to display sequence and structure information. Bioinformatics, 18, 767–768. [DOI] [PubMed] [Google Scholar]
- 9.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brünger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S. et al. (2000) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 12.Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graphics, 15, 132–134. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst., 24, 946–950. [Google Scholar]
- 14.Servant F., Bru,C., Carrère,S., Courcelle,E., Gouzy,J., Peyruc,D. and Kahn,D. (2002) ProDom: automated clustering of homologous domains. Brief. Bioinf., 3, 246–251. [DOI] [PubMed] [Google Scholar]
- 15.Rost B. (1996) PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- 16.Combet C., Blanchet,C., Geourjon,C. and Deléage,G. (2000) NPS@: network protein sequence analysis. Trends Biochem. Sci., 25, 147–150. [DOI] [PubMed] [Google Scholar]
- 17.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henrissat B. and Bairoch,A. (1996) Updating the sequence-based classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J., 293, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robert X. (2002) PhD thesis, Université Claude Bernard Lyon I.
- 20.Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) CATH—a hierarchic classification of protein domain structures. Structure, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- 21.Notredame C., Higgins,D.G. and Heringa,J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]