Abstract
A number of biological data resources (i.e. databases and data analytical tools) are searchable and usable on-line thanks to the internet and the World Wide Web (WWW) servers. The output from the web server is easy for us to browse. However, it is laborious and sometimes impossible for us to write a computer program that finds a useful data resource, sends a proper query and processes the output. It is a serious obstacle to the integration of distributed heterogeneous data resources. To solve the issue, we have implemented a SOAP (Simple Object Access Protocol) server and web services that provide a program-friendly interface. The web services are accessible at http://www.xml.nig.ac.jp/.
INTRODUCTION
The International Nucleotide Sequence Database (INSD) is composed of DNA Data Bank of Japan (DDBJ), EMBL Nucleotide Database at the European Bioinformatics Institute and GenBank at the National Center for Biotechnology Information in US and has accumulated nucleotide sequences and their biological meaning (annotation). The size of INSD exceeded 20 million entries and 20 billion nucleotides in 2002 (http://www.ddbj.nig.ac.jp/ddbjnew/statistics-e.html). Data processing by hand is not feasible any more. In addition to the explosion of the INSD, data from genome, proteome, transcriptome, physiome and other comprehensive studies on life phenomena wait to be analyzed. The integration of these databases and data analysis tools is indispensable in bioinformatics. However, the integration is a laborious task because most data resources have been developed ad hoc. In addition, the data structure and the syntax of query and result are often not transparent.
In the case of the INSD, the three data banks exchange data everyday in a consistent data structure that is composed of features, qualifiers and values (http://www.ddbj.nig.ac.jp/sub/ref1-e.html). The INSD provides the data to the public also in the same data structure as used in the exchange. The most popular INSD data format visible to anonymous users is the so-called flat file format (FF format). Therefore, it is possible to write a script to parse the FF format. It is ironical that the quite similar script for a slightly different aim has been repeatedly written by a number of groups in the world. ‘There is massive duplication of efforts’ (1) in parsing many other web pages too.
It is also to be noted that the INSD has modified and expanded the features and qualifiers in accordance with the progress of biotechnology and life sciences (i.e. the FF format has changed). Thus a script that parses FF format has to be updated by referring to the document provided by the INSD (http://www.ddbj.nig.ac.jp/sub/ref1-e.html) sometimes. Nevertheless, the INSD may claim that it provides comparatively stable data structure and format. There are many other biological data sources that are valuable but do not provide a sufficient document for users and change the access method and the data format.
Thus it keeps bioinformaticians busy to analyze multiple data sources and update scripts. It will not be feasible to integrate data from diverse data sources in this way in the near future, because a tsunami of data will be diffused by a number of research groups in addition to public data banks. There are two avenues to save the situation.
One avenue is standardization. The Inter-Union Bioinformatics Group [a Joint Initiative of the International Union for Pure and Applied Biophysics (IUPAB), the International Union of Biochemistry and Molecular Biology (IUBMB), the International Union of Crystallography (IUCr), the International Union of Pure and Applied Chemistry (IUPAC) and the Committee on Data for Science and Technology (CODATA)] pointed out in the report (http://www.wdcm.org/iubg.html) the needs for:
standardization of data definitions and nomenclature; and
standardization of data formats and data exchange;
for the improvement of ‘the availability, maintenance, and free access of biological and biophysical scientific data’ (http://www.wdcm.org/iubg.html). It will make the life of bioinformaticians easy and more productive, if every data source follows a guideline on standardization. The INSD set of the features, qualifiers and values can be a model for the first standardization. The FF format is a base for the second standardization and the INSD in eXtensible Markup Language (XML) document (2) may be the model. Although we have to pursue the standardization, there is a long way to go to prepare standards for every aspect of life phenomena and make the standards accepted by all the data providers.
The other avenue is to improve the interoperability of data resources while the specification of each data resource remains as it is. We tested Common Request Broker Architecture (CORBA) for the interoperability (3,4) because we supposed that CORBA is an interoperable open architecture. After the evaluation of CORBA and other methods, we selected and introduced SOAP (http://www.w3.org/TR/SOAP/) and Web Services Description Language (WSDL) (http://www.w3.org/TR/wsdl) into our site.
WHY SOAP?
We applied CORBA to an e-Workbench (3) that realized a seamless and reversible operation to capture, store, retrieve and analyze phenotypic and sequence data of microbes. We also applied CORBA to a distributed database system in the Genome Information Broker (4). In the meantime, we experienced that CORBA is really a powerful tool to integrate distributed databases and application software in a local area network (LAN). European Bioinformatics Institute that is the host of EMBL Nucleotide Database is another active institute in providing CORBA servers to the public (http://corba.ebi.ac.uk/). However, we also recognized that CORBA is not feasible in practice for the integration of multiple servers distributed in the wide area network (WAN), even if servers can interface with CORBA. CORBA is built on the Internet Inter-Object Request Broker (ORB) Protocol (IIOP) but IIOP is firewall-unfriendly. The firewall does not transmit IIOP in the default setting. In addition, all of system managers in outside institutes refused to let IIOP tunnel through firewall when we approached them to extend the e-Workbench from LAN to WAN. Furthermore, we experienced that ORBs developed by different tools were actually incompatible. Analysis of each ORB and a custom program for each combination of ORBs are required to make ORBs interoperable.
SOAP uses XML documents for message passing so that it is firewall-friendly in contrast with IIOP. In addition, SOAP is independent from platform and computer language. Therefore, we can expect that SOAP servers and clients are interoperable even in WANs. Let us demonstrate by a simple use case that SOAP servers greatly simplify the integration of multiple data sources compared to either by web surfing or by CGI programs. The use case is: I want to retrieve an INSD entry that was referred to in a published paper, match the entry to records in multiple databases by BLAST homology search and get results in a unified format. The procedures of each mode will be as follows.
- Web surfing:
- type the accession number in a web browser connected to the INSD site;
- edit the result from the INSD to get the sequence data;
- execute BLAST against the databases A, B, C and so on;
- edit results from the multiple databases;
- store the edited data into a private file or database.
- CGI program:
- analyze a method and structure of queries to the INSD and multiple databases;
- analyze the data structure and format of results of the INSD and the multiple databases;
- develop a script or program to create and send a query and process the result for each server;
- trace URL addresses and check multiple servers in order to modify scripts when necessary.
- SOAP server:
- call appropriate method provided by SOAP servers.
You will be able to properly use bio-* such as bioperl, bioJava, bioruby and biopython to access biological databases, if you know the specification of the database such as the scheme, output format and so on. By use of our SOAP server, the user will be able to more directly access biological objects from the INSD without worrying about the change of the data format and items. It is one step closer to the user than the bio-* when the INSD is concerned.
SOAP SERVER PREPARED BY US
Our SOAP server is implemented into the hardware of Pentium III 1.3 GHz×2, 4 Gb memory and 90 Gb hard disk. The set of software is RedHat Linux 7.2J, Jakarta Tomcat 4.0.3, Xerces 1.4.4, Ant 1.2, GLUE 1.3, PostgreSQL 7.2.1, Java (J2SE 1.3.1), BioJava 1.21, Java Mail API 1.2 and Java Activation Framework 1.0.1. The SOAP server provides the following services:
GetEntry: get entries by specifying accession numbers;
BLAST homology search;
FASTA homology search;
Smith–Waterman homology search;
ClustalW: multiple alignment;
SRS: Sequence Retrieval System;
TxSearch: retrieval of scientific names and lineage of an organism;
DDBJ: retrieve the DDBJ entries and extract some features.
Users have been able to browse the result of these services using a web browser and download by email server and FTP server. After the implementation of the SOAP server, any user can call the service in the above from his/her program in JAVA or Perl. A sample program to call a method in the SOAP server for GetEntry is:
#!/usr/bin/perl
use SOAP::Lite;
my $service=SOAP::Lite-
>service(‘http://xml.nig.ac.jp/wsdl/GetEntry.wsdl’);
$result=$service->getXML_DDBJEntry (“AB000003”) ;
print $result;
Other examples of methods and results are introduced in Tables 1 and 2.
Table 1. An example of the method of the web services named DDBJ: retrieve biological features (annotation) that correspond to sub-sequences in a specified region (between bases 59 000 and 64 000) of the entry of AL121903.
| Method | getRelatedFeatures(“AL121903”,“59000”,“64000”) |
| Result | repeat_region 60476..60773 |
| CDS join(59036..59120,63774..63916) | |
| repeat_region 60170..60462 | |
| mRNA join(59025..59120,63774..63916) | |
| source 59000..64000 | |
| misc_feature join(59025..59120,63774..63822) | |
| repeat_region 60774..61063 | |
| repeat_region 61066..61361 | |
| repeat_region 62066..62373 | |
| repeat_region 61740..62041 | |
| CDS join(59036..59120,63774..63916) | |
| repeat_region 63994..64000 | |
| mRNA join(59025..59120,63774..63916) | |
| repeat_region 59422..59716 | |
| repeat_region 63193..63243 |
Table 2. An example of combining two methods of the web services named BLAST: carry out homology search by BLASTP and then identify the ranges that the query sequence and an entry sequence in the INSD match (align) in the result of BLASTP. The result is transferred from the first method to the second method by the parameter ‘&result’.
| Methods | search(“blastp”, “SWISS”, “MSSRIARALALVVTLLHLTRLALSTCPAACHCPLEAPKCAPGVGLVRDGCGCCKVCAKQL”) | ||
| extractPosition($result) | |||
| A part of result | sp|O00622|CYR6_HUMAN | ||
| Query | 1 | 60 | |
| Hit | 1 | 60 | |
| sp|Q9ES72|CYR6_RAT | |||
| Query | 24 | 60 | |
| Hit | 24 | 60 | |
| sp|P18406|CYR6_MOUSE | |||
| Query | 24 | 60 | |
| Hit | 24 | 60 | |
| sp|P19336|CE10_CHICK | |||
| Query | 24 | 60 | |
| Hit | 24 | 60 | |
| sp|Q9Z0G4|CTGL_MOUSE | |||
| Query | 26 | 60 | |
| Hit | 26 | 60 | |
| sp|O18739|CTGF_BOVIN | |||
| Query | 26 | 60 | |
| Hit | 29 | 64 | |
The system requirement of the SOAP client is GLUE or Apache Axis library in the case of call from Java. In the case of call from Perl, SOAP-Lite, libwww-perl, MIME-Base64, URI and XML-Parser are required.
XML Central OF DDBJ
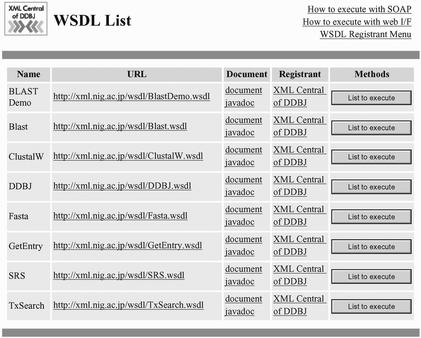
The SOAP server is actually accessible at the HomePage of XML Central of DDBJ (http://xml.nig.ac.jp) that is introduced in Figure 1. You might still suspect that CGIs are easier to utilize than understanding the SOAP server and methods. To make you overcome the barrier, we prepared a user-friendly interface to access the SOAP server by use of the Web Service Description Language (WSDL). You are able to try the SOAP server without writing a program in this environment. If you click the menu of ‘Web services’ in Figure 1, you get the list of web services as shown in Figure 2. You will find out the method, if you click the ‘Execute’ button, as follows:
search(program, database, query, param);
search(program, database, query);
extractPosition(result);
Figure 1.
HomePage of XML Central of DDBJ at http://www.xml.nig.ac.jp/.
Figure 2.
The web services available from http://www.xml.nig.ac.jp/. Click ‘List to execute’ and get the list of methods prepared for each web service.
for the method of BLAST.
If you are interested in the first method, just click it to get the interactive interface as shown in Figure 3. You may be able to understand the function of the SOAP server through this interface and write your program.
Figure 3.
The interactive interface to utilize a method of ‘getRelatedFeatures(accession, start, stop)’ in the web service of ‘DDBJ’. The result in shown in Table 1.
As you might recognize in Figure 1, you can not only use our SOAP server but also register your SOAP servers to XMLCentral of DDBJ. Our site is in the public domain and the registration will benefit bioinformaticians both in academia and industries all over the world.
CONCLUDING REMARKS
You will recognize that XML technology prevails in biology, if you search the internet by ‘XML’ and ‘bio’. However, diverse DTD and/or XML schemes are proposed for the same object. In addition, a number of computer programs have been written to process data in a semi-structured text file such as the flat file. Therefore, we prepare output of the web services both in a simple text file and an XML document. We also prepare a web interface for novice users to understand a method. Therefore our site will be complementing other frameworks of the web services such as BioMOBY (5) by lowering the barrier to the web services.
We expect that the world of the web services will expand and the directory of the web services will be interoperable and comprehensive including the directory provided by BioMOBY. Then it will be quick and easy for bioinformaticians to locate and call the service from their system, even though biological data resources keep expanding. Rich work flows for specific research aims will be also available in the public domain, if the web services world become more mature. We will report sample work flows elsewhere.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mr Yasumasa Shigemoto and Mr Mashito Yamaguchi of Fujitsu for their support in the development of SOAP servers and WSDL. We are also grateful to the Ministry of Education, Science, Culture, Sports and Technology and also Japan Science and Technology Corporation for their enduring financial support.
REFERENCES
- 1.Stein L. (2002) Creating a bioinformatics nation. Nature, 417, 119–120. [DOI] [PubMed] [Google Scholar]
- 2.Miyazaki S., Sugawara,H., Gojobori,T. and Tateno,Y. (2003) DNA Data Bank of Japan (DDBJ) in XML. Nucleic Acids Res., 31, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugawara H. and Miyazaki,S. (2001) An integrated retrieval and analysis system for microbial data distributed in the Internet. InfroBIO. American Society for Microbiology, May 20–24, Orlando, USA. [Google Scholar]
- 4.Fumoto M., Miyazaki,S. and Sugawara,H. (2002) Genome Information Broker (GIB): data retrieval and comparative analysis system for completed microbial genome and more. Nucleic Acids Res., 30, 66–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilkinson Mark D. and Links,M. (2002) BioMOBY: an open source biological web services proposal. Brief. Bioinf., 3, 331–341. [DOI] [PubMed] [Google Scholar]