Abstract
The WWW servers at http://www.icgeb.trieste.it/dna/ are dedicated to the analysis of user-submitted DNA sequences; plot.it creates parametric plots of 45 physicochemical, as well as statistical, parameters; bend.it calculates DNA curvature according to various methods. Both programs provide 1D as well as 2D plots that allow localisation of peculiar segments within the query. The server model.it creates 3D models of canonical or bent DNA starting from sequence data and presents the results in the form of a standard PDB file, directly viewable on the user's PC using any molecule manipulation program. The recently established introns server allows statistical evaluation of introns in various taxonomic groups and the comparison of taxonomic groups in terms of length, base composition, intron type etc. The options include the analysis of splice sites and a probability test for exon-shuffling.
INTRODUCTION
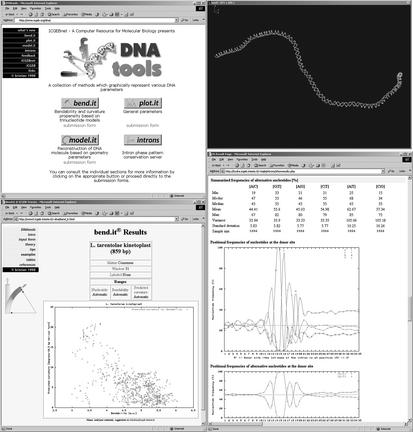
The thinking of biologists has been profoundly influenced by the idea of local structural polymorphism in DNA. DNA is no longer considered as a featureless polymer but rather as a series of individual domains differing in flexibility and curvature. In addition to helical polymorphism (B, A or Z structures), here we often deal with a localised micropolymorphism in which the original B-DNA structure is only distorted but is not extensively modified (1). Visualisation is an essential task as the underlying features can be identified neither by inspecting the sequences nor by looking for specific sequence patterns. The DNA tools page incorporates a number of visualisation tools that produce 1D, 2D and 3D representations in response to DNA sequence queries as well as a newly established site for intron statistics (Fig. 1).
Figure 1.
The DNA tools server. Bottom left: results of a 2D plot, curvature versus DNA bendability plot in Leishmania tarentolae kinetoplast DNA (EMBL: AF380674) produced by bend.it. Top right: 3D model of a 400 bp segment of DNA, calculated by model.it and displayed by Swiss-PDBviewer (9). Bottom right: positional frequency plot of nucleotides at donor splice site in plant DNA (3556 non/redundant plant introns), output of the IS server.
THE SERVERS
plot.it
The plot.it server produces parametric plots using various physicochemical parameters (2). A query sequence is divided into overlapping nmers and the average value of a given parameter is calculated using tabulated values. The results appear either as simple sequence plots or as 2D plots in which two parameters are plotted against each other.
bend.it
The bend.it server calculates the curvature of DNA molecules as predicted from the DNA sequences. The calculation is based on values tabulated for dinucleotides and trinucleotides, and the curvature (degree per helical turn) is calculated using standard algorithms (1). This calculation was originally based on DNA bendability parameters derived from DNAseI digestion that characterise the (static or dynamic) bending of trinucleotides towards the major groove (3). Today a number of other dinucleotide (4–6) and trinucleotide models (3,7) are included, and the results can be visualised as 1D or 2D plots on the screen. Both the bend.it and the plot.it servers are based on C programs provided with GnuPlot graphic routines. Their output appears on the screen and is optionally sent by email to the user.
model.it
The model.it server was designed to provide 3D models of DNA in response to DNA sequence queries (8). The results are presented as a standard PDB file that can be viewed directly using any of the widely available molecule manipulation programs such as Swiss-PDBviewer (9) or RasMol (10). In addition to straight A and B DNA models, the server is capable of building curved DNA models using the parameter sets mentioned above. The server program was written using NAB—a high level molecule manipulation language (11). Coordinates of the sugar–phosphate backbone are optionally optimised with constrained molecular dynamics using energy parameters from the AMBER package (12). At present, the server can produce models of 700 bp in length, but models >50 bp will not be optimised. Modelling of canonical, straight B or A DNA structures proceeds in a similar way, but without the need for backbone geometry optimisation.
IS introns
The IS introns server was designed to provide statistical overviews on intron groups (13). Simple questions, like comparison of introns between various taxonomic groups in terms of intron phases or size distributions as well as the analysis of splice sites, requires a carefully selected dataset as well as meticulous work that has to be repeated as new data become available. The goal of the introns server was to esatablish an automatically updated intron resource that allows the evaluation of experimentally validated and statistically balanced intron datasets, as well as a flexible comparison of groups according to various criteria. In addition to sequence retrieval and BLAST similarity search, there are options to compare taxonomic groups based on the NCBI Taxonomy Database, and to perform on-the-fly statistics. The analysis capabilities include statistical evaluation (minimum, maximum, average, standard deviation, etc.) of intron and exon length, of the number of introns per gene, base composition, intron phases, as well as a graphic comparison of two or more groups in terms of the above variables. In addition, the analysis of splice sites and testing of the exon shuffling hypothesis (14,15) are explicitly included.
CONCLUSION
All the servers are provided with help files that describe the detailed instructions, the theory, the literature citations as well as the instructions for installing the accessory programs such as Swiss-PDBviewer (9) or RasMol (10).
Acknowledgments
ACKNOWLEDGEMENTS
The development of the web servers was partly funded by EMBnet, the European Molecular Biology Network in the framework of EU project QLRI-CT-2001-01363 and by the EU-project ORIEL (IST-2001-32688) coordinated by the European Molecular Biology Organisation (EMBO).
REFERENCES
- 1.Munteanu M.G., Vlahovicek,K., Parthasarathy,S., Simon,I. and Pongor,S. (1998) Rod models of DNA: sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem. Sci., 23, 341–347. [DOI] [PubMed] [Google Scholar]
- 2.Vlahovicek K., Gabrielian,A. and Pongor,S. (1998) Prediction of bendability and curvature in genomic DNA. J. Math. Model. Scient. Comput., 9, 53–57. [Google Scholar]
- 3.Brukner I., Sanchez,R., Suck,D. and Pongor,S. (1995) Trinucleotide models for DNA bending propensity: comparison of models based on DNaseI digestion and nucleosome packaging data. J. Biomol. Struct. Dyn., 13, 309–317. [DOI] [PubMed] [Google Scholar]
- 4.Ulyanov N.B. and James,T.L. (1995) Statistical analysis of DNA duplex structural features. Methods Enzymol., 261, 90–120. [DOI] [PubMed] [Google Scholar]
- 5.Bolshoy A., McNamara,P., Harrington,R.E. and Trifonov,E.N. (1991) Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl Acad. Sci. USA, 88, 2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olson W.K., Marky,N.L., Jernigan,R.L. and Zhurkin,V.B. (1993) Influence of fluctuations on DNA curvature. A comparison of flexible and static wedge models of intrinsically bent DNA. J. Mol. Biol., 232, 530–554. [DOI] [PubMed] [Google Scholar]
- 7.Gabrielian A. and Pongor,S. (1996) Correlation of intrinsic DNA curvature with DNA property periodicity. FEBS Lett., 393, 65–68. [DOI] [PubMed] [Google Scholar]
- 8.Vlahovicek K. and Pongor,S. (2000) Model.it: building three dimensional DNA models from sequence data. Bioinformatics, 16, 1044–1045. [DOI] [PubMed] [Google Scholar]
- 9.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 10.Sayle R.A. and Milner-White,E.J. (1995) RasMol: biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]
- 11.Macke T. and Case,D.A. (1998) In Leontes,N.B. and SantaLucia,J. (eds), Molecular Modeling of Nucleic Acids. American Chemical Society, Washington DC, pp. 379–393. [Google Scholar]
- 12.Case D.A., Pearlman,D.A., Caldwell,J.W.,III, T.E.C., Ross,W.S., Simmerling,C.L., Darden,T.A., Merz,K.M., Stanton,R.V., Cheng,A.L. et al. (1997) AMBER 5. 5th edn. University of California, San Francisco. [Google Scholar]
- 13.Barta E., Kajan,L. and Pongor,S. (2003) IS: a web-site for intron statistics. Bioinformatics, 19, 543. [DOI] [PubMed] [Google Scholar]
- 14.Long M., de Souza,S.J. and Gilbert,W. (1995) Evolution of the intron-exon structure of eukaryotic genes. Curr. Opin. Genet. Dev., 5, 774–778. [DOI] [PubMed] [Google Scholar]
- 15.Kriventseva E.V. and Gelfand,M.S. (1999) Statistical analysis of the exon-intron structure of higher and lower eukaryote genes. J. Biomol. Struct. Dyn., 17, 281–288. [DOI] [PubMed] [Google Scholar]